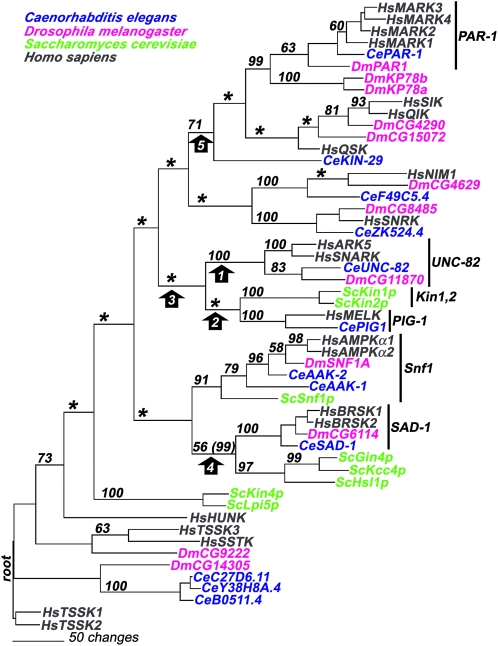
Figure 7.—
The parsimony tree presents a model of the descent of UNC-82 and related kinases from a common ancestral sequence. The tree was rooted to the HsTSSK sequences as shown in the human kinome tree (Manning et al. 2002; http://www.kinase.com). Bootstrap values (out of 100) are indicated on each branch; asterisks denote nodes that did not receive support >50. The proposed orthology of the UNC-82/ARK5/SNARK proteins is well supported (bootstrap value 100) for the node that separates this group from all other sequences (arrow 1). In contrast, neither the node connecting UNC-82 to the Kin1 and PIG-1 groups (arrow 2) nor the node that separates the UNC-82, Kin1, and PIG-1 groups from all other groups (arrow 3) is supported by bootstrap analysis. Two groupings that receive moderate bootstrap support in this parsimony analysis (arrows 4 and 5) receive higher support (in parentheses) using neighbor-joining techniques: the node grouping the SAD-1 family with the trio of yeast kinases Gin4/Kcc4/Hsl1 and the node grouping the PAR-1 family with HsSIK, HsQSK, and CeKIN-29.