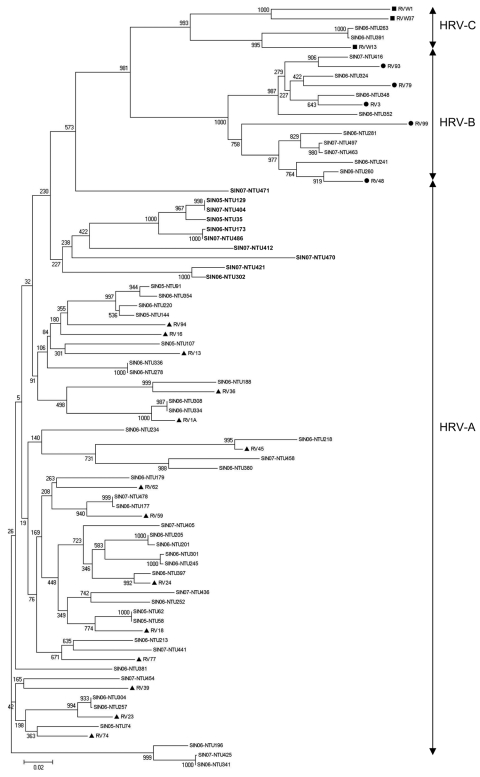
Figure.
Phylogenetic analysis of human rhinoviruses (HRVs) from Singapore based on nucleotide sequences of the 5′ noncoding region. The tree was constructed by using the neighbor-joining method with 1,000 bootstrapped replicates generated by MEGA version 4 software (9). Sequences (GenBank accession nos. FJ645828–FJ645771) of viruses from Singapore (SIN) are indicated, where the 2 numbers represent the year the specimen was collected, and NTU (Nanyang Technological University) followed by 3 numbers represents the specimen number. Representative strains of HRV-C are indicated by squares, HRV-B by circles, and HRV-A by triangles. RV indicates rhinovirus strains, followed by the serotype no. These sequences were obtained from the report by Lee et al. (3). Boldface indicates a cluster of 10 HRV-A strains that diverged from reference HRV-A strains. Scale bar indicates nucleotide substitutions per site.