Abstract
The profiling of subproteomes from complex mixtures on the basis of small molecule interactions shared by members of protein families or small molecule interaction domains present in a subset of proteins is an increasingly important approach in functional proteomics. Capture CompoundTM Mass Spectrometry (CCMS) is a novel technology to address this issue. CCs are trifunctional molecules that accomplish the reversible binding of target protein families to a selectivity group (small molecule), covalent capturing of the bound proteins by photoactivated cross-linking through a reactivity group, and pullout of the small molecule-protein complexes through a sorting function, e.g. biotin. Here we present the design, synthesis, and application of a new Capture Compound to target and identify cAMP-binding proteins in complex protein mixtures. Starting with modest amounts of total protein mixture (65–500 μg), we demonstrate that the cAMP-CCs can be used to isolate bona fide cAMP-binding proteins from lysates of Escherichia coli, mammalian HepG2 cells, and subcellular fractions of mammalian brain, respectively. The identified proteins captured by the cAMP-CCs range from soluble cAMP-binding proteins, such as the catabolite gene activator protein from E. coli and regulatory subunits of protein kinase A from mammalian systems, to cAMP-activated potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channels from neuronal membranes and specifically synaptosomal fractions from rat brain. The latter group of proteins has never been identified before in any small molecule protein interaction and mass spectrometry-based proteomics study. Given the modest amount of protein input required, we expect that CCMS using the cAMP-CCs provides a unique tool for profiling cAMP-binding proteins from proteome samples of limited abundance, such as tissue biopsies.
cAMP is an important biological second messenger molecule involved in many biological processes, such as adaptation of bacteria to low glucose growing conditions, chemotaxis in slime molds, and various signal transduction processes in metazoa downstream of the activation of hormone receptors (1). The concentration level of cAMP in biological systems is tightly controlled by the activity of adenylyl cyclases that catalyze the formation of cAMP and by the activity of phosphodiesterases, which catalyze the degradation of cAMP. Given the importance of signaling cascades downstream of hormone or neurotransmitter receptors that involve increased formation or degradation of cAMP, the identification and profiling of cAMP effector proteins can be expected to be an essential contribution to elucidate the molecular basis of physiological as well as pathophysiological signaling events.
Bona fide effectors of cAMP are proteins that contain a cyclic nucleotide binding domain (CNBD).1 This motif represents a protein domain initially defined and characterized by the crystal structure of the major known cAMP-binding protein from Escherichia coli, the catabolite gene activator protein (2). This domain is present in all known mammalian cAMP-binding proteins as well. Three major classes of proteins exist that contain CNBDs. The first group contains protein kinase A subunits, namely regulatory subunits of protein kinase A isozymes (3), as well as the cGMP-dependent protein kinases (4). A group of Rap guanine nucleotide exchange factors (Epac proteins) that contain CNBDs (5) comprises the second group. Both groups contain key proteins involved in signaling cascades. A number of ion channels that can be directly regulated by cAMP contain CNBDs, such as the cyclic nucleotide-gated channels (6), make up the third group. In particular, potassium/sodium hyperpolarization-activated cyclic nucleotide-gated (HCN) channels play a crucial role in the pacemaking of heart and brain activity (7). A relatively small number of further proteins that contain CNBDs, such as phosphodiesterase isoforms and a sodium-hydrogen exchange transporter, can be retrieved from searches in databases such as Swiss-Prot.
Among the methodological repertoire applied in functional proteomics, small molecule affinity-based techniques seem to be ideal for the task of profiling the cAMP binding proteome subset. Established strategies make use of cAMP affinity beads. These beads comprise cAMP derivatives covalently attached to the polymer backbone via an aminoalkyl linker. The linker may vary in length of the alkyl chain and in the attachment position at the nucleobase (8, 9). This approach, however, suffers from the relatively large amount of protein input required to obtain significant data, precluding e.g. the profiling of the target proteins in samples of limited abundance. Furthermore, it has not been demonstrated yet that affinity-based enrichment of cAMP-binding proteins is suitable for cAMP-binding membrane proteins that are known to be difficult to access. On the other hand, soluble cAMP- and cGMP-binding proteins along with their interaction partners were robustly identified with this methodology. Another approach described in the literature used a cyclic guanosine monophosphate analogue immobilized on a Biacore chip to isolate cGMP- and cAMP-binding proteins from a cell lysate, estimate the quantity of the material, and elute proteins for proteolysis and identification by LC-MS/MS. In addition, for single purified proteins, binding constants can be measured (10). The applicability of this approach to transmembrane cGMP/cAMP-binding proteins, however, has yet to be determined.
Here we describe the synthesis and application of a trifunctional Capture CompoundTM (CC) (see Fig. 1A) as a novel approach for the functional isolation of cAMP-binding proteins from complex protein mixtures using low amounts of protein input. In contrast to current pulldown approaches, the CC enables the covalent linkage to the target proteins by a photoactivatable reactivity group in addition to the reversible binding of target proteins by the selectivity group. The Capture Compound-protein conjugate can be isolated from the complex protein mixture via the sorting function (a biotin moiety) of the Capture Compound by means of streptavidin-coated magnetic beads (see Fig. 1, B and C) (11). The cAMP-binding protein-selective Capture Compound described here was successfully applied to the isolation of cAMP-binding proteins from E. coli lysate and cultured eukaryotic HepG2 cells, respectively. Furthermore, we report the applicability of the CCMS approach for the capturing of cAMP-binding HCN channel proteins from rat brain synaptosome preparations as well. To our knowledge, this has not yet been achieved by any cAMP affinity bead approach. In addition, the ion channels, which by antibody- and in situ hybridization-based techniques have been shown to be located in neuronal tissues at synaptic sites (12, 13), have also escaped detection in many detailed proteomics profiling studies conducted to establish the protein complements of synaptic structures (see e.g. Refs. 14–17). Our data suggest that the cAMP-CC approach is uniquely efficient and sensitive for the identification and profiling of cAMP-binding proteins in complex protein mixtures.
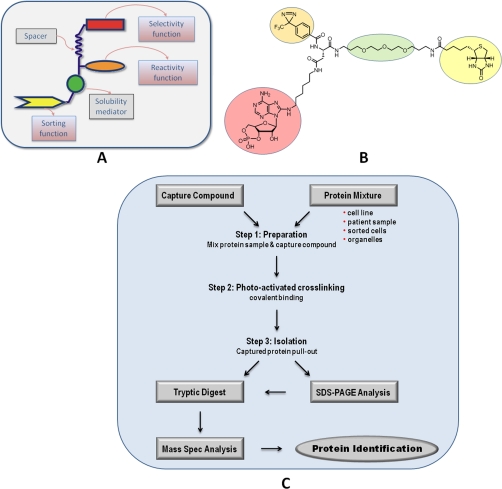
Fig. 1.
A, schematic design of a CC. Three functionalities are coupled to a core. The selectivity function (red), e.g. modified cAMP, for target recognition; the reactivity function (orange), e.g. diazirines, for covalent cross-linking; and the sorting function (yellow), e.g. biotin, for pullout of captured proteins; and a variable linker (green) that can modify the hydrophilicity of the system are shown. B, structure of 8-AHA-cAMP-CC (7c), which represents one of several cAMP-CCs that are available. C, flow chart of the CCMS technology.
EXPERIMENTAL PROCEDURES
Materials
All standard chemical reagents and solvents were obtained from commercial suppliers and were used without further purification. Compound 1 (the scaffold) was synthesized according to the developed route, and compounds 6-AE-cAMP (2a), 6-AH-cAMP (2b), 2-AEA-cAMP (3a), 2-AHA-cAMP (3b), 8-AEA-cAMP (4a), 8-ABA-cAMP (4b), 8-AHA-cAMP (4c), and 8-ADOA-cAMP (4d) were purchased from Biolog Life Science Institute (Bremen, Germany). Flash chromatography was performed with silica gel (0.035–0.070 mm; Acros Oganics, Geel, Belgium), and reaction progress was determined by thin-layer chromatography (ethyl acetate/methanol = 1:1) using Merck silica gel plates at 254 nm UV light or by visualization using appropriate reagents and heating of the silica gel plate. Medium pressure LC purification was performed on a Büchi Sepacore system (equipped with Büchi C-605 pumps and UV photometer C-635 detector) using a mobile phase of methanol/water with 0.1% acetic acid at a flow rate of 15 ml/min, products were purified on a Lichroprep RP Select B (25–40 μm; Merck) 15 × 200-mm column. 1H NMR, 13C NMR spectra were recorded using a Bruker Avance 400 spectrometer in MeOH-d4 or DMSO-d6. The purity of the compounds was determined using a Waters LCT ToF MS system (equipped with a Waters 1525 pump and a Waters 2998 UV photometer) using a mobile phase of ACN/water with 0.1% formic acid at a flow rate of 0.4 ml/min; the column applied was a Phenomenex Synergi Fusion RP (2.5 μm, 20 × 2 mm). The capture experiments were performed using caproBoxTM and caproMagTM (caprotec bioanalytics GmbH, Berlin, Germany) according to the manufacturer's instructions. E. coli lysate was a kind gift from E. Weinhold (Rheinisch-Westfälische Techische Hochschule (RWTH), Aachen, Germany), and HepG2 cell lysate was purchased from InVivo (Berlin, Germany). Deep frozen rat brain was purchased from BLS Preclinical Services (Berlin, Germany). MS grade ACN, methanol, ethanol, and water were purchased from Merck; trypsin was purchased from Roche Applied Science; and bovine protein kinase A, regulatory subunit type II (PKARII) was purchased from Calbiochem (Lot number B74298). Human protein kinase A, regulatory subunit type Iα (PKARIα) was purchased from Biaffin (Kassel, Germany).
Chemical Synthesis of cAMP-CCs: General Procedure
To a solution of scaffold 1 (10 μmol) in N,N-dimethylacetamide (1 ml) was added 4-dimethylaminopyridine (25 μmol), N,N′-diisopropylcarbodiimide (DIC) (50 μmol), N-hydroxybenzotriazole (HOBt) (30 μmol), and the relevant cAMP derivative (2a, 2b, 3a, 3b, 4a, 4b, 4c, or 4d; 5 μmol) under argon. The reaction mixture was stirred at 50 °C under argon. After 16 h, the solvent was removed under vacuum, and the residue obtained was purified by medium pressure LC using methanol and 0.1% acetic acid to afford the relevant cAMP-CCs (6-AE-cAMP-CC (5a), 6-AH-cAMP-CC (5b), 2-AEA-cAMP-CC (6a), 2-AHA-cAMP-CC (6b), 8-AEA-cAMP-CC (7a), 8-ABA-cAMP-CC (7b), 8-AHA-cAMP-CC (7c), and 8-ADOA-cAMP-CC (7d)). For detailed analytical data, see the supplemental material.
Stability of cAMP-CCs and Competitors
Chemical, UV, and biological stability of cAMP-CCs were tested prior to usage in CCMS experiments. For the testing of the chemical stability, 1 ml of cAMP-CC (30 μm) was incubated at 80 °C for 30 min in a heating block. After cooling to room temperature, the compound was mixed with 10 μl of the internal standard reserpine (200 μg/ml) and measured directly by HPLC-MS as described above. For UV stability testing, 100 μl of cAMP-CC (30 μm) were irradiated in the caproBox at 4 °C for 20 min. Subsequently, the compound was mixed with 3 μl of the internal standard and measured directly by HPLC-MS. The biological stability of cAMP-CCs and the corresponding competitors was evaluated in E. coli lysate (40 mg/ml) at 37 °C, in HepG2 lysate (8 mg/ml) at 30 and 4 °C, and in 3′,5′-cyclic phosphodiesterase (PDE; 0.5 units; Sigma) at 30 °C. In short, 50 μl of cAMP-CC (100 μm) were mixed with E. coli lysate (500 μg of protein), HepG2 lysate (500 μg of protein), or PDE (0.5 units), respectively, in the presence of buffer. At defined time points, 100 μl of the incubation mixture were taken and mixed with 500 μl of cold 2-propanol (−20 °C) for overnight protein precipitation at −20 °C. After centrifugation at 15,000 rpm (21,000 × g) for 30 min at 4 °C in a table top centrifuge (Universal 320R centrifuge, Hettich, Kirchlengern, Germany), the supernatant was dried under vacuum, and the residue was redissolved in 200 μl of water. After addition of 3 μl of internal standard, HPLC-MS measurements were carried out (see the supplemental material).
Measurement of EC50 of 8-AHA-cAMP-CC (7c) Using Fluorescence Polarization
The fluorescence polarization competitive binding experiment was performed at the department of biochemistry, University of Kassel, Kassel, Germany, following the procedure from Moll et al. (18). For further details, see the supplemental material. The EC50 of competitor 8-AHA-cAMP (4c) has been determined previously (18).
Capturing in E. coli and HepG2 Lysate and in Rat Brain Synaptosomal Fractions
The capture experiments were performed as follows. For on-bead experiments, 25 μl of cAMP-CC (100 μm) were incubated with 50 μl of Dynabeads® MyOneTM Streptavidin C1 (Invitrogen Dynal) in a 0.2-ml PCR tube with flat cap (Thermo Scientific) at room temperature for 30 min. Afterward, the beads were collected in the cap of the PCR tube using the caproMag, a magnet device developed for the convenient collection of magnetic beads in up to 12 parallel samples (caprotec bioanalytics GmbH), and washed twice with wash buffer (WB) containing 50 mm Tris-HCl (pH 7.5), 1 mm EDTA, 1 m NaCl, 0.05% (w/v) n-octyl-β-glucopyranoside, according to the caproMag user instructions. The beads were carefully resuspended in a PCR tube containing 100 μl of the analyzing mixture. This mixture comprises 20 μl of 5× concentrated capture buffer (100 mm HEPES, 250 mm KOAc, 50 mm Mg(OAc)2, 50% glycerol, 1% (w/v) Triton X-100 (pH 7.5)) and 500 μg of cell lysate (E. coli lysate or HepG2 lysate). The reaction mixture was rotated at 4 °C for ∼3 h under protection from light. For photocross-linking, the reaction mixtures were irradiated in closed PCR tubes using the caproBox for 20 min in 2.5-min intervals and shaking the tubes in between (it is important to note that magnetic beads must not remain in the tube cap). The characteristics of the caproBox (see the supplemental material) were as follows: capture temperature, 0.5–4 °C; λmax, 312 nm (for a detailed spectrum of the lamp, see the supplemental material); irradiance Ie, 10–12 milliwatts/cm2; irradiation energy for each sample at 15-mm2 irradiation area of the closed tube and 10-mm height of the reaction mixture, ∼1.4 J. After UV light exposure, the beads were collected using the caproMag and washed six times with 200 μl of WB and twice with water. After washing, the beads were finally collected in the cap and were ready for the subsequent processing step.
For off-bead experiments, 10 μl of cAMP-CCs (100 μm) were mixed with 500 μg of cell lysate and 20 μl of 5× concentrated capture buffer in a 0.2-ml PCR tube with flat cap (Thermo Scientific). The mixture was incubated at 4 °C for 30 min and subsequently irradiated in open PCR tubes for 10 min using the caproBox. The characteristics of the caproBox were the same as described for on-bead experiments except the irradiation area (23-mm2 irradiation area when open tubes were used). The irradiation energy applied to each sample was ∼1.5 J. The samples were mixed with 25 μl of 5× concentrated WB. Then 50 μl of Dynabeads MyOne Streptavidin C1 beads were added to the reaction mixture. Additional incubation at 4 °C for 30 min was carried out under rotation. The beads were collected using the caproMag according to the user instructions and washed six times with 200 μl of WB and twice with water. The beads were then ready for further processing.
The capture experiments in the rat brain synaptosomal fractions preparations (for preparation, see below) were performed similarly to the capturing in E. coli and HepG2 lysate although with slight modifications with respect to protein concentration and WB: ∼65 μg of protein were applied in capture experiments, and 0.1% n-dodecyl-β-maltoside (Glycon, Luckenwalde, Germany) was added to the capture and the wash buffer.
Preparation of Rat Brain Synaptosomes
Synaptosomes are artificial organelle-like structures that form upon mild homogenization of neuronal tissue. They contain the former presynaptic terminals of neurons with material of the postsynaptic structures of the former postsynaptic neurons attached (19). Rat brain synaptosomal fractions were isolated essentially according to tom Dieck et al. (20). Briefly, two rat brains were homogenized in a motor-driven glass-Teflon homogenizer (Sartorius, Goettingen, Germany) in 10 volumes (as compared with tissue wet weight) of homogenization buffer (0.32 m sucrose, 5 mm HEPES/NaOH (pH 7.4), Complete EDTA-free protease inhibitor cocktailTM (Roche Applied Science)) using 12 strokes at 900 rpm at 4 °C. The homogenate was centrifuged at 1000 × g (Hettich) for 10 min. The supernatant was decanted and further centrifuged at 12,000 × g for 20 min. The supernatant (S2) from this procedure was snap frozen, and the protein concentration was determined according to Bradford (21). The pellet (P2) was resuspended in 0.32 m sucrose (30× weight of brain in grams) and further homogenized in a motor-driven glass-Teflon homogenizer using six strokes at 900 rpm at 4 °C. The suspension was centrifuged at 12,000 × g for 20 min. The pellets (P2′) from this procedure were resuspended in 0.32 m sucrose, Tris-HCl (5 mm, pH 8.1) using six strokes at 900 rpm and carefully placed on top of a 2.5-ml sucrose density gradient solution (bottom layer, 1.2 m sucrose; middle layer, 1.0 m sucrose; top layer, 0.8 m sucrose; volume of the layers, 3.5 ml, respectively). Ultracentrifugation was performed at 85,000 × g for 120 min (WX-80 Ultra, rotor TH641, Thermo Scientific).
After centrifugation the upper layer (0.32 m/0.8 m interphase) contained myelin, one middle layer (0.8 m/1.0 m interphase) contained light membranes, the other middle layer (1.0 m/1.2 m interphase) contained synaptosomal fractions, and the bottom layer contained mitochondria (1.2 m/pellet).
The synaptosomal fractions were solubilized using cell opening buffer (6.7 mm MES, 6.7 mm NaOAc, 6.7 mm HEPES (pH 7.6), 200 mm NaCl, 10 mm β-mercaptoethanol) with protease inhibitor (Roche Applied Science) and 0.5% n-dodecyl-β-maltoside. The suspension was rotated at 900 rpm for 1 h at room temperature and centrifuged at 10,000 × g for 15 min at 4 °C. The supernatant was collected, and the protein concentration was determined according to Bradford (21). The supernatant was used for capture experiments.
Protein Digestion
Following the capture experiments, two different protocols for protein digestion were applied.
In-solution Digestion (Protein Digestion Directly on the Magnetic Bead Particles)
After protein capturing, the streptavidin magnetic beads were washed twice with 200 μl of LC-MS grade water. For tryptic digestion, the streptavidin magnetic bead protein mixture was incubated together with 9 μl of 50 mm ammonium bicarbonate, 0.5 mm calcium chloride and 1 μl of trypsin (0.5 μg/ml) (Roche Applied Science) for 16 h at 37 °C on a temperature-controlled shaker. Subsequently, tryptic peptides were desalted using Stage tips® (Proxeon Biosystems A/S, Odense, Denmark) and eluted according to the manufacturer's instructions. The eluate was evaporated to dryness in a miVac DNA vacuum centrifuge (Genevac®, Suffolk, UK) and stored at −20 °C until mass spectrometric analysis.
In-gel Digestion
After protein capturing, streptavidin magnetic beads were heated to 95 °C for 10 min in Laemmli sample buffer (22) and applied to gel electrophoresis. Subsequently, proteins in the SDS gel were stained using the Proteo Silver staining kit (Sigma) according to the manufacturer's instructions. After the staining, the gel was washed twice for 10 min with LC-MS grade water. Gel bands were excised, cut in small pieces, and washed twice with 100 μl of water and 100 μl of 50% ethanol (v/v). For shrinking, each gel band was incubated with 50 μl of 100% ethanol for ∼5 min. Subsequently, the washing and shrinking steps were repeated. For protein digestion, the gel pieces were rehydrated in a solution of 12.5 ng/μl trypsin, 50 mm ammonium bicarbonate incubated for 45 min on ice. After replacing the supernatant with 5–20 μl of 50 mm ammonium bicarbonate without trypsin, the samples were incubated for 16 h at 37 °C. The extraction of peptides was done in two consecutive steps by incubating the gel pieces with 50% ACN, 2.5% formic acid (FA) for 15 min. The pooled supernatants were then dried in a miVac DNA vacuum centrifuge. Desalting, elution, evaporation, and storage of tryptic peptides were performed as described for “in-solution digestion” samples.
Nano-LC-MS/MS
The protein digest was redissolved in 5 μl of 0.1% FA and sonicated briefly. Subsequently, peptides were loaded directly onto a nanoflow Biosphere C18 precolumn (5 μm, 120 Å, 20 × 0.1 mm; NanoSeparations, Nieuwkoop, Netherlands) coupled to a nanoflow Biosphere C18 analytical column (5 μm, 120 Å, 105 × 0.075 mm). The experiments were performed on an Easy-nLCTM liquid chromatography system (Proxeon Biosystems A/S) connected to an LTQ Orbitrap XL mass spectrometer (Thermo Electron, Bremen, Germany) utilizing a nanoelectrospray ion source (Proxeon Biosystems A/S).
For the analysis of in-solution digest samples, peptides were eluted during an 80-min linear gradient from 5% ACN, 0.1% FA to 40% ACN, 0.1% FA followed by an additional 2 min to 100% ACN, 0.1% FA and remaining at 100% for another 8 min with a controlled flow rate of 300 nl/min. For the analysis of extracted gel bands, a linear 40-min gradient increasing from 5% ACN, 0.1% FA to 40% ACN, 0.1% FA was used followed by an additional 2 min to 100% ACN, 0.1% FA and remaining at 100% for another 8 min with a controlled flow rate of 300 nl/min.
The mass spectrometric analysis was performed in the data-dependent mode to automatically switch between orbitrap-MS and LTQ-MS/MS (MS2) acquisition. The mass spectrometer duty cycle was controlled by setting the injection time automatic gain control. Survey full-scan MS spectra (from m/z 400 to 2000) were acquired in the orbitrap with resolution r = 60,000 at m/z 400 (after accumulation to a target value of 500,000 charges in the linear ion trap). The most intense ions (up to five, depending on signal intensity) were sequentially isolated for fragmentation in the linear ion trap using CID at a target value of 10,000 charges. The resulting fragment ions were recorded in the LTQ.
For accurate mass measurements in the MS mode, the singly charged polydimethylcyclosiloxane background ion (Si(CH3)2O)6H+ (m/z 445.120025) generated during the electrospray process from ambient air was used as the lock mass for real time internal recalibration. Target ions already mass-selected for CID were dynamically excluded for the duration of 60 s. Charge state screening and rejection of ions for CID with unassigned charge were set. Further mass spectrometric settings were as follows: spray voltage was set to 1.7 kV, temperature of the heated transfer capillary was set to 200 °C, and normalized collision energy was 35% for MS2. The minimal signal required for MS2 was 500 counts. An activation q = 0.25 and an activation time of 30 ms were applied for MS2 acquisitions. After each analysis of an in-solution digest sample, the system was washed by performing at least one linear gradient that was used for the respective peptide separation.
Peptide Identification via Database Search
All MS/MS samples were analyzed using SEQUEST implemented in BioworksBrowser 3.3.1 SP1 (Thermo Fisher Scientific) and X!Tandem (The Global Proteome Machine Organization; version 2007.01.01.1). Automated database searching against the human, E. coli, and rat UniProtKB/Swiss-Prot database (release 56.6, containing 410,518 sequence entries) was performed with 5-ppm precursor tolerance, 1-amu fragment ion tolerance, and full trypsin specificity allowing for up to two missed cleavages. Phosphorylation at serine, threonine, and tyrosine; oxidation of methionines; deamidation at asparagines and glutamine; acetylation at lysine and serine; formylation at lysine; and methylation at arginine, lysine, serine, threonine, and asparagine were allowed as variable modifications. No fixed modifications were used in the database search.
Probability assessment of peptide assignments and protein identifications were made through the use of Scaffold 2 (version Scaffold_2_04_00, Proteome Software Inc., Portland, OR) by combining SEQUEST and X!Tandem database searches. Only peptides with ≥95% probability as specified by the Peptide Prophet algorithm (23) were considered. Protein identification probabilities for multiple peptide assignments were set to ≥95% according to the Protein Prophet algorithm (24). Proteins that comprise similar peptides and could not be differentiated based on MS/MS analysis alone were grouped to satisfy the principles of parsimony. Single peptide protein identifications were only accepted if the peptide probability was ≥95%. Furthermore, these protein hits were manually validated by inspection of the MS/MS spectra (see the supplemental material).
Protein identifications were only accepted and reported as distinct hits if they were based on at least one discriminant (unique) peptide that passed probability validation, according to Scaffold analysis. The estimated false discovery rate of peptide identifications was determined using the reversed protein database approach and was <0.5%.
RESULTS
Chemistry and Stability Characteristics of cAMP-CCs
Capture Compounds are trifunctional small organic molecules that can be used for equilibrium binding of target proteins to the selectivity group, subsequent covalent capture of the target protein via a photoactivatable reactivity group, and pullout of the Capture Compound-protein conjugates via a sorting function, e.g. biotin. The schematic design of the Capture Compounds (A), the structure of a cAMP-CC (B), and the general capture protocol (C) are depicted in Fig. 1.
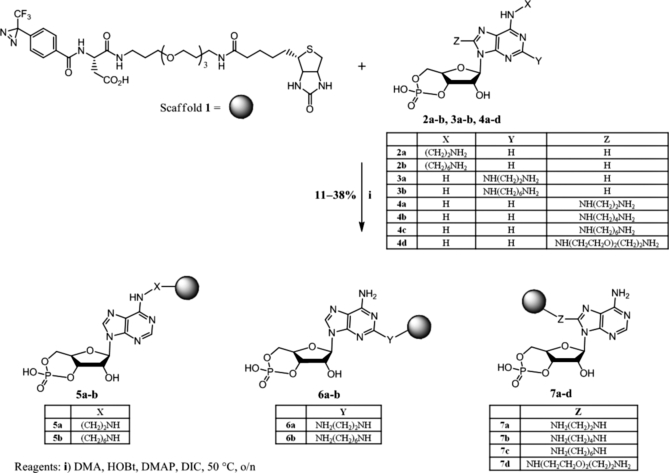
We synthesized a series of cAMP-CCs, which possess different attachment sites at the cAMP selectivity group linked to the Capture Compound scaffold 1. The series of cAMP-CCs was synthesized because we expected different binding properties to the target protein families with differently attached cAMP-CCs. The linker used to attach the selectivity group to scaffold 1 varied in its chain lengths and in its properties (hydrophobic versus hydrophilic). In total, eight different cAMP-CCs were synthesized that covered the C2, C8, and N6 position of the adenine moiety. The linker length of the cAMP-CCs ranged from two (ethyl) to eight (3,6-dioxaoctyl) atoms. The synthetic route to scaffold 1 will be published elsewhere.2 Coupling of scaffold 1 and the aminoalkyl-modified cAMP derivatives (2a, 2b, 3a, 3b, 4a, 4b, 4c, and 4d) was carried out using a standard condensing method (DIC/HOBt), providing the desired cAMP-CCs (5a, 5b, 6a, 6b, 7a, 7b, 7c, and 7d) in satisfying yields (11–38%, not optimized; Scheme 1). All eight cAMP-CCs were stable against heat and UV irradiation (with respect to the non-photoactivatable moiety of the compounds) and stable in E. coli lysate (40 mg of protein/ml) at 37 °C as well. The biological stability of Capture Compound 7c and the corresponding precursor 4c (which should also be used as competitor) was further explored in HepG2 lysate (8 mg of protein/ml) at 4 and 30 °C; the latter compound was also explored in the presence of PDE at 30 °C. Generally, Capture Compound 7c as well as competitor 4c was stable in HepG2 cell lysate at 4 °C. Precursor 4c was also stable in the presence of PDE for 2 h. The formation of very small amounts of 8-AHA-AMP-CC (from 7c) or 8-AHA-AMP (from 4c) was only observed at prolonged incubation times (at least 180 min in HepG2 cell lysate at 4 °C or with PDE at 30 °C). However, in HepG2 cell lysate at 30 °C, substantial decomposition of the starting materials was observed within 180 min (see the supplemental material). No difference in stability was found for Capture Compounds that comprise other attachment position at the cAMP moiety (e.g. cAMP-CC 5b; see the supplemental material).
Scheme 1.
Chemical synthesis of the eight cAMP-CCs. The gray ball refers to scaffold 1. DMA, N,N-dimethylacetamide; DMAP, 4-dimethylaminopyridine; o/n, overnight.
Different Coupling Positions and Linker Lengths of the cAMP-CCs Display Different Capturing Characteristics in E. coli Lysate
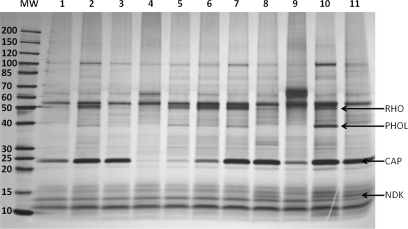
We chose lysate from the E. coli K12 strain DH5α to test the functionality of our Capture Compounds on bona fide cAMP-binding proteins. E. coli contains one bona fide cAMP-binding protein, the catabolite gene activator protein (CAP), which upon cAMP binding induces a change in gene expression in response to altered nutritional conditions (26, 27). All eight cAMP-CCs synthesized were tested in capture experiments using E. coli lysate (500 μg of total protein). The capture experiments were performed on bead and off bead, respectively (for details, see “Experimental Procedures”). All cAMP-CCs captured CAP (Table I). Nevertheless, the capture efficiency differed significantly depending on the linker length. In a comparative study for the capture efficiency of C8-attached cAMP-CCs with different linker lengths (7a, 7b, 7c, and 7d) (Fig. 2, lanes 1–4), we observed that capturing of CAP was significantly lower with 8-AEA-cAMP-CC (7a) than with the other Capture Compounds (7b, 7c, and 7d). This was confirmed by subsequent nano-LC-MS/MS analysis regarding both the number of identified peptides and the number of assigned spectra (Table I). In the case of 7a, only five CAP-specific peptides were identified in the on-bead capture experiment, whereas just one CAP-specific peptide was identified in the off-bead experiment (see Table I). In general, the longer the linker, the better the cAMP-CC binds to CAP with an apparent optimum for the hexyl linkers as judged on the basis of protein band intensities on the gel and the number of peptides (assigned spectra) identified in LC-MS/MS. However, when the linker length was too long, the capture efficiency decreased again as shown with compound 7d. Capturing of CAP using 8-AHA-cAMP-CC (7c) was largely reduced in the presence of an excess amount of 2 mm 8-AHA-cAMP (4c) as competitor (Fig. 2, lane 5), whereas 2 mm cAMP as competitor was less efficient (Fig. 2, lane 6) because of the low stability of cAMP in E. coli lysate. Addition of scaffold 1 to 8-AHA-cAMP-CC (7c) did not influence the capture yield (Fig. 2, lane 7), providing additional evidence that capturing of CAP is solely based on the interaction between the selectivity function and the target protein. The competition efficiency (Table I, last column) differed significantly between off-bead (two identified peptides) and on-bead (six identified peptides) experiments. One reason might be that in contrast to off-bead experiments on-bead experiments were carried out with a 2.5 times higher concentration of Capture Compound. This is necessary because of the streptavidin magnetic beads, which were also used in 2.5-fold excess in both cases but were fully loaded with Capture Compound in the on-bead experiment. The changed ratio between Capture Compound and competitor to a less optimal value may cause incomplete competition. Another explanation for the reduced competition efficiency of 8-AHA-cAMP (4c) in on-bead experiments might be substantial steric hindrance at the protein-coated surface of the beads that disturbed the thermodynamic equilibrium. Comparing 6-AH-cAMP-CC (5b) and 6-AE-cAMP-CC (5a) as well as 2-AHA-cAMP-CC (6b) and 2-AEA-cAMP-CC (6a) (Table I and Fig. 2, lanes 8–11) again revealed that CCs with a longer linker have a better capture efficiency, although the difference was less pronounced in the case of 6a versus 6b. The tendency for a C4–C6 linker is in line with two requirements for the Capture Compound design: 1) the “undisturbed” binding of the selectivity group must be preserved, and 2) the reactivity group must be able to cross-link to the target protein. Based on their ability to capture CAP, the three hexyl-linked cAMP-CCs, 8-AHA-cAMP-CC (7c), 6-AH-cAMP-CC (5b), and 2-AHA-cAMP-CC (6b), were further explored in several comparative experiments. However, the present work is focused on a single cAMP-CC, compound 7c, although the capability to capture cAMP-binding proteins has been demonstrated for all other cAMP-CCs and will be reported in detail in due course.3
Table I. Identified peptides and assigned spectra (in parentheses) of the CAP after cAMP-CC capturing in E. coli lysate (500 μg of total protein).
Scaffold 1 was used as the background control. No CAP was captured with this compound.
| 8-ADOA-cAMP-CC (7d) | 8-AHA-cAMP-CC (7c) | 8-ABA-cAMP-CC (7b) | 8-AEA-cAMP-CC (7a) | 6-AH-cAMP-CC (5b) | 6-AE-cAMP-CC (5a) | 2-AHA-cAMP-CC (6b) | 2-AEA-cAMP-CC (6a) | 8-AHA-cAMP Cmptna | |
|---|---|---|---|---|---|---|---|---|---|
| Off bead | 7 (7) | 9 (24) | 10 (13) | 1 (1) | 11 (46) | 10 (20) | 11 (44) | 8 (9) | 2 (2) |
| On bead | 12 (25) | 13 (56) | 13 (27) | 5 (5) | 12 (58) | 12 (16) | 11 (87) | 13 (60) | 6 (9) |
a 8-AHA-cAMP Cmptn refers to competition experiments using Capture Compound 7c in the presence of 2 mm 8-AHA-cAMP (4c).
Fig. 2.
Silver-stained SDS-PAGE gel (4–20% gradient gel): on-bead capturing profiles in E. coli lysate (500 μg) using eight different cAMP-CCs. Lane 1, 8-ADOA-cAMP-CC (7d); lane 2, 8-AHA-cAMP-CC (7c); lane 3, 8-ABA-cAMP-CC (7b); lane 4, 8-AEA-cAMP-CC (7a); lane 5, competition of 7c with 2 mm 8-AHA-cAMP (4c); lane 6, competition of 7c with 2 mm cAMP; lane 7, 1:1 mixture of 7c and scaffold 1; lane 8, 6-AH-cAMP-CC (5b); lane 9, 6-AE-cAMP-CC (5a); lane 10, 2-AHA-cAMP-CC (6b); lane 11, 2-AEA-cAMP-CC (6a). MW, molecular weight marker; NDK, nucleoside-diphosphate kinase; PHOL, PhoH-like protein; rho, transcription termination factor Rho.
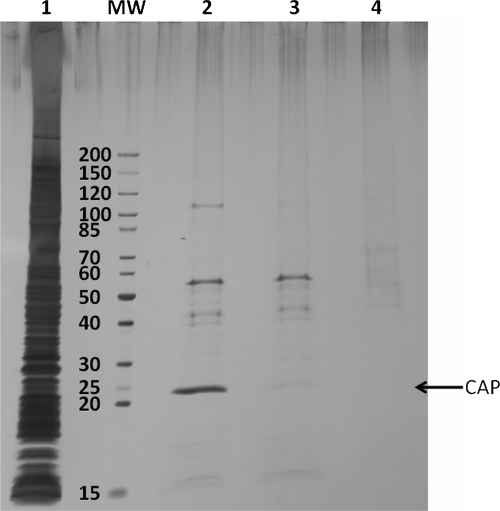
We found that besides cAMP-dependent proteins several other nucleotide-binding proteins were specifically captured although in low amounts (as judged by SDS-PAGE) and identified with a low number of unique peptides. In addition, each cAMP-CC captured a certain subset of these nucleotide-binding proteins (data will be published in due course).3 For instance, the 8-AHA-cAMP-CC (7c) is the only Capture Compound that specifically captured glutamate 5-kinase (ProB) as identified by LC-MS/MS (in the gel (see Fig. 2, lane 2), glutamate 5-kinase and PhoH-like protein (PHOL) co-migrated due to their very similar molecular masses, 39,057 versus 39,039 Da). On the other hand, 6-AH-cAMP-CC (5b), 2-AHA-cAMP-CC (6b), and 2-AEA-cAMP-CC (6a) exclusively captured the nucleoside-diphosphate kinase (Fig. 2, lanes 8, 10, and 11, NDK). Some other proteins, such as PhoH-like protein and transcription termination factor rho (RHO), were captured only by compounds 6b and 7c, although they were not completely absent in competition experiments using either cAMP or 8-AHA-cAMP (4c). However, scaffold 1 did not capture these proteins. The reason might be a much higher binding affinity of the Capture Compounds in contrast to the (modified) nucleotides. The enrichment efficiency for Capture Compound-binding proteins was further demonstrated in a case study where CAP was isolated from E. coli cell lysate (Fig. 3). In this experiment, the capturing (photocross-linking, lane 2) was compared with (a) the competition using 2 mm 8-AHA-cAMP (4c) (lane 3) and (b) a pulldown experiment (without photocross-linking, lane 4). From Fig. 3, it can be clearly stated that the simple pulldown experiment is much less effective to enrich CAP than the capture experiment according to our protocol.
Fig. 3.
Silver-stained SDS-PAGE gel (4–20% gradient gel): on-bead capturing in E. coli lysate (500 μg) using Capture Compound 7c. Lane 1, 1 μg of E. coli whole cell lysate; lane 2, 8-AHA-cAMP-CC (7c) with photoactivation (=capturing); lane 3, competition of 7c with 2 mm 8-AHA-cAMP (4c); lane 4, 8-AHA-cAMP-CC (7c) without photoactivation (=pulldown). MW, molecular weight marker.
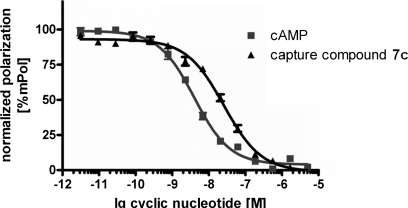
We extended our studies to mammalian cAMP-binding proteins. In an initial experiment, we compared the EC50 value of Capture Compound 7c with that of cAMP to cAMP-binding proteins, such as the commercially available regulatory subunits of PKA, by fluorescence polarization. In the fluorescence polarization competitive binding experiment (28), the binding potency of the cyclic nucleotides was directly related to the obtained EC50 value as shown in Fig. 4. It was found that cAMP displays an EC50 value of 4 nm and the Capture Compound 7c displays an EC50 value of 25.8 nm, indicating a 6 times decreased affinity of the Capture Compound to human PKARIα, although the Capture Compound 7c is still a nanomolar binder to this protein. Based on the binding assay and the capture results from E. coli, compounds 5b, 6b, and 7c along with 8-AEA-cAMP-CC (7a) were tested using a commercially available preparation of the bovine PKARII. Again, the regulatory subunit of PKA bound cAMP with nanomolar affinity. As expected, the C6-linked 8-AHA-cAMP-CC (7c) displayed a much better capture performance than the ethyl-linked analog (7a) (Fig. 5, lanes 5 and 7). The two other hexyl-linked Capture Compounds with N6 and C2 attachment positions, 5b and 6b, were similarly efficient compared with Capture Compound 7c. In analogy to the experiments with the E. coli lysate, the addition of 2 mm cAMP to the reaction prior to the addition of the CCs largely abolished capturing (Fig. 5, lanes 2, 4, 6, and 8). Scaffold 1, which lacks the selectivity function, showed no significant binding to bovine PKARII (Fig. 5, lane 9). This result enabled us to use the scaffold 1 as a second background control. Because the gel bands obtained from capture experiments using 2.3 μg of bovine PKARII with Capture Compounds 5b, 6b, and 7c (Fig. 5, lanes 1, 3, and 5) were of similar intensities compared with the control band comprising 0.5 μg of bovine PKARII (lane 10), a capture yield of ∼20% was estimated with all of these Capture Compounds. It should be noted that photocross-linking yields are often below 10% (29), so a capture yield of over 20% is remarkable. The capture yield of 7c was independently determined to be 23% by analyzing an SDS-PAGE gel where a triplicate capture experiment was compared with a concentration series of bovine PKARII (0.1–0.5 μg) (see supplemental Fig. F12). In an analogous experiment, the capture yield for human PKARIα with Capture Compound 7c was determined (37%), providing insight into the variability of the photocross-linking efficiency even for closely related, but distinct proteins.
Fig. 4.
Fluorescence polarization competitive binding experiment. 2.5 nm human PKARIα was added to a serial dilution of the cyclic nucleotide indicated on the plot, and apparent EC50 values were calculated (for experimental details see the supplemental material). Each data point represents the mean ± SEM (standard error of the mean) from nine individual wells from three different experiments.
Fig. 5.
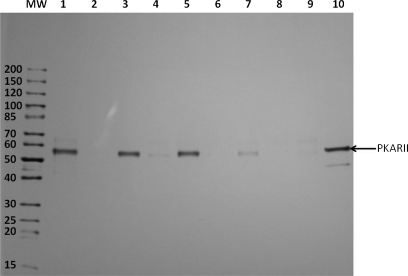
Silver-stained SDS-PAGE gel (4–20%): on-bead capturing of purified bovine PKARII (2.3 μg) using four different cAMP-CCs and scaffold 1 as control. Lane 1, 2-AHA-cAMP-CC (6b); lane 2, competition with cAMP (2 mm); lane 3, 6-AH-cAMP-CC (5b); lane 4, competition with cAMP (2 mm); lane 5, 8-AHA-cAMP-CC (7c); lane 6, competition with cAMP (2 mm); lane 7, 8-AEA-cAMP-CC (7a); lane 8, competition with cAMP (2 mm); lane 9, scaffold 1; lane 10, pure PKARII (0.5 μg). MW, molecular weight marker.
Capturing cAMP-binding Proteins from HepG2 Cell Lysate
The 8-AHA-cAMP-CC (7c) was chosen to perform capture experiments in more complex mammalian systems. In these experiments, 500 μg of the human hepatocyte-derived cell line HepG2 lysate were used for each capture reaction. The major proteins identified in these capture experiments by LC-MS/MS were endogenous regulatory subunits of the cAMP-dependent protein kinase (KAP) (Table II). Notably, capturing of these proteins was completely abolished in the presence of 8-AHA-cAMP (4c) (2 mm), which acted as a competitor (Table II). Interestingly, protein kinase A-anchoring proteins (AKAP1 and AKAP9), which are known to be bona fide interaction partners of the regulatory subunits of PKA, were also identified.
Table II. Identified cAMP-binding proteins and the protein kinase A-anchoring proteins in capture experiments using 8-AHA-cAMP-CC (7c) in HepG2 cell lysate (500 μg of total protein).
No proteins were captured in competition experiments when capturing was carried out in the presence of 2 mm 8-AHA-cAMP (4c). Proteins captured by scaffold 1 were disregarded except for AKAP1, which showed binding to 1 although with a low number of identified peptides and assigned spectra. The identified unique peptides have a probability of ≥95% (for peptide sequences see supplemental material).
| No. | Protein name | UniProt/Swiss-Prot accession number | No. of identified peptides (runs 1–3) | No. of assigned spectra (runs 1–3) | Coverage | Molecular mass |
|---|---|---|---|---|---|---|
| % | kDa | |||||
| 1a | KAP0, cAMP-dependent protein kinase type I-α regulatory subunit | P10644 | 12/16/8 | 19/25/8 | 32/34/32 | 43 |
| 2a | KAP1, cAMP-dependent protein kinase type I-β regulatory subunit | P31321 | 1/2/0 | 1/2/0 | 4/5/0 | 43 |
| 3 | KAP2, cAMP-dependent protein kinase type II-α regulatory subunit | P13861 | 15/18/8 | 22/37/13 | 43/56/27 | 46 |
| 4b | AKAP1, mitochondrial | Q9Y2D5 | 5/6/0 | 5/6/0 | 7/8/0 | 97 |
| 5 | AKAP9 | Q99996 | 1/4/0 | 1/4/0 | 0.2/1.1/0 | 454 |
a Some peptides identified for KAP0 (P10644) also match to KAP1 (P31321) (see supplemental material).
b AKAP2 (Q92667) was also identified in two runs, although each identification was based on only one peptide (peptide probability, 95%; protein probability, 70%; sequence coverage, 1.1%; see supplemental material).
In addition to the known cAMP-binding proteins, several further proteins with no reported cAMP binding activity were identified in these CCMS experiments (Table III). With a few exceptions (RL18A, RL6, and GNL3), these proteins were not detected in competition experiments using 8-AHA-cAMP (4c) (2 mm). Because most of these proteins contain a nucleotide binding domain, competition experiments were (also) successful using other nucleotides (2 mm) as competitors, such as ATP, GTP, NAD, or NADH (see Table III).
Table III. Identified nucleotide-binding proteins in capture experiments using 8-AHA-cAMP-CC (7c) in HepG2 cell lysate (500 μg of total protein).
The proteins were almost completely abolished (not more than one assigned spectrum in total) in competition experiments when capturing was carried out in the presence of 2 mm 8-AHA-cAMP (4c). Proteins captured by scaffold 1 were disregarded. The identified unique peptides have a probability of ≥95% (sequences are reported in the supplemental material).
| No. | Protein name | UniProt/Swiss-Prot accession number | No. of identified peptides (runs 1–3) | No. of assigned spectra (runs 1–3) | Coverage | Alternative competitiona | Molecular mass |
|---|---|---|---|---|---|---|---|
| % | kDa | ||||||
| 1b | RL6, 60 S ribosomal protein L6 | Q02878 | 5/4/3 | 6/4/3 | 19/18/13 | NAD, (ATP) | 33 |
| 2b | RL18A, 60 S ribosomal protein L18a | Q02543 | 7/7/2 | 7/7/3 | 29/32/9 | NADH, (GTP) | 21 |
| 3 | NCPR, NADPH-cytochrome P450 reductase | P16435 | 3/3/2 | 3/4/3 | 6/8/7 | (GTP) | 77 |
| 4b | GNL3, guanine nucleotide-binding protein-like 3 | Q9BVP2 | 6/4/1 | 6/5/1 | 15/11/4 | GTP | 62 |
| 5 | LU, Lutheran blood group glycoprotein | P50895 | 5/4/1 | 5/4/1 | 11/8/1.8 | ATP | 67 |
| 6 | M6PBP, mannose 6-phosphate receptor-binding protein 1 | O60664 | 4/3/1 | 4/3/1 | 13/10/4 | NADH | 47 |
| 7 | VDAC2, voltage-dependent anion-selective channel protein 2 | P45880 | 2/4/1 | 2/4/1 | 8/15/3 | NAD | 32 |
| 8 | IMMT, mitochondrial inner membrane protein | Q16891 | 4/2/2 | 4/2/2 | 6/3/7 | NADH | 84 |
| 9 | TXTP, tricarboxylate transport protein, mitochondrial | P53007 | 3/5/0 | 5/5/0 | 11/18/0 | NAD | 34 |
| 10 | NDUS3, NADH dehydrogenase (ubiquinone) iron-sulfur protein 3, mitochondrial | O75489 | 2/3/2 | 2/3/2 | 11/15/12 | NADH | 30 |
| 11 | SSRD, translocon-associated protein subunit δ | P51571 | 3/4/1 | 3/4/1 | 20/31/11 | NADH | 19 |
| 12 | STML2, stomatin-like protein 2 | Q9UJZ1 | 4/5/1 | 4/5/1 | 16/21/4 | NADH | 39 |
| 13 | NDK8, putative nucleoside-diphosphate kinase | O60361 | 1/3/1 | 1/3/1 | 12/34/7 | ATP | 16 |
a Nucleotides in parentheses did not give complete competition. Incomplete competition was defined to be the case when two or more assigned spectra but less than the half of those found in the capture experiments were identified in all competition experiments.
b Competition failed with RL6, RL18a, and GNL3. However, competition was possible in the presence of 2 mm NAD (RL6), NADH (RL18A), and GTP (GNL3), respectively.
Capturing Proteins from Rat Brain Synaptosomal Fractions
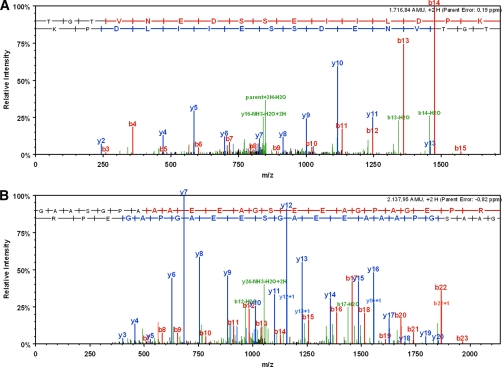
To assess the capability of cAMP-CCs to isolate other classes of cAMP-binding proteins, we sought to perform the capture experiment in rat brain synaptosomal fractions. Synaptosomes from rat brain were prepared according to standard procedures (20) (see “Experimental Procedures”). Starting with 65 μg of total protein mixture (from the solubilized synaptosomes), we were able to isolate and identify three cAMP-dependent protein kinases, one cGMP-dependent protein kinase, two AKAPs, and further bona fide cAMP-binding proteins using 8-AHA-cAMP-CC (7c). Among the bona fide cAMP-binding proteins were two members of the HCN channels (Fig. 6, A and B, for MS/MS spectra) and a Rap guanine nucleotide exchange factor (Table IV). In the control experiment using 8-AHA-cAMP (4c) (2 mm) as competitor, no cAMP-binding protein-specific peptides were identified by LC-MS/MS. In addition to proteins with known CNBD and their binding partners, other nucleotide-binding proteins were captured with 7c as well but not with scaffold 1 (Table V). In contrast to the experiments with HepG2 lysate, competition experiments using 2 mm 8-AHA-cAMP (4c) failed with synaptosomal fractions. However, competition (fully or in part) was possible with other nucleotides (2 mm concentration of either ATP, GTP, or NADH; Table V). Only the mitochondrial phosphate carrier protein (MPCP) was not efficiently abolished with any (cyclic) nucleotide as competitor.
Fig. 6.
A, MS/MS spectrum of HCN1_RAT potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 1-specific tryptic peptide TGTVNEDSSEIILDPK (Swiss-Prot accession number Q9JKB0). B, MS/MS spectrum of HCN2_RAT potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel 2-specific tryptic peptide GAASGPAAAEEAGSEEAGPAGEPR (Swiss-Prot accession number Q9JKA9).
Table IV. Identified cAMP-binding proteins and the protein kinase A-anchoring proteins in capture experiments using 8-AHA-cAMP-CC (7c) in rat brain synaptosomal fractions (65–80 μg of total protein mixture as input).
All proteins, except the cGMP-depending protein kinase KGP2, were almost completely abolished in competition experiments when capturing was carried out in the presence of 2 mm 8-AHA-cAMP (4c) or 2 mm ATP, respectively. Proteins captured by scaffold 1 were disregarded. The identified unique peptides have a probability of ≥95% (sequences are reported in the supplemental material).
| No. | Protein name | UniProt/Swiss-Prot accession number | No. of identified peptides (runs 1–4) | No. of assigned spectra (runs 1–4) | Coverage | Molecular mass |
|---|---|---|---|---|---|---|
| % | kDa | |||||
| 1 | KAP0, cAMP-dependent protein kinase type I-α regulatory subunit | P09456 | 10/5/1/5 | 13/7/1/6 | 30/13/2/13 | 43 |
| 2 | KAP2, cAMP-dependent protein kinase type II-α regulatory subunit | P12368 | 17/16/10/11 | 29/26/12/13 | 37/30/26/30 | 46 |
| 3 | KAP3, cAMP-dependent protein kinase type II-β regulatory subunit | P12369 | 12/14/6/9 | 27/32/8/17 | 34/37/21/35 | 46 |
| 4 | AKAP5 | P24587 | 10/17/4/2 | 11/19/4/2 | 18/26/7/5 | 76 |
| 5 | AKAP18 δ isoform (protein kinase A (PRKA)-anchoring protein 7) | Q6JP77 | 3/3/1/1 | 3/4/1/1 | 9/6/3/3 | 39 |
| 6 | KGP2, cGMP-dependent protein kinase 2 | Q64595 | 5/10/3/2 | 5/11/3/2 | 9/15/5/3 | 87 |
| 7 | HCN1 | Q9JKB0 | 6/8/5/2 | 7/10/5/2 | 12/15/12/5 | 102 |
| 8a | HCN2 | Q9JKA9 | 8/11/5/3 | 9/11/5/3 | 16/19/7/5 | 95 |
| 9 | RPGF4, Rap guanine nucleotide exchange factor 4 | Q9Z1C7 | 1/2/0/1 | 1/2/0/1 | 3/5/0/3 | 50 |
a HCN3 (Q9JKA8) was also identified in two of four runs, although each identification was based on only one peptide (peptide probability, 95%; protein probability, 58 and 75%; sequence coverage, 1.8 and 2%; see supplemental material).
Table V. Identified nucleotide-binding proteins in capture experiments using 8-AHA-cAMP-CC (7c) in rat brain synaptosomal fractions (65–80 μg of total protein mixture as input).
All proteins were captured even in competition experiments when capturing was carried out in the presence of 2 mm 8-AHA-cAMP (4c). However, competition experiments were successful with other nucleotides (see column “Competition”). Proteins captured by scaffold 1 were disregarded except for DHE3, which showed binding to 1 although with a low number of identified peptides and assigned spectra. The identified unique peptides have a probability of ≥95% (sequences are reported in the supplemental material).
| No. | Protein name | UniProt/Swiss-Prot accession number | No. of identified peptides (runs 1–4) | No. of assigned spectra (runs 1–4) | Coverage | Competitiona | Molecular mass |
|---|---|---|---|---|---|---|---|
| % | kDa | ||||||
| 1 | DHE3, glutamate dehydrogenase 1, mitochondrial | P10860 | 14/27/22/14 | 17/47/36/20 | 28/44/35/32 | GTP, NADH | 61 |
| 2 | IMMT, inner membrane protein, mitochondrial | Q3KR86 | 13/14/4/8 | 15/16/5/8 | 27/30/8/19 | (ATP, GTP) | 67 |
| 3b | MPCP | P16036 | 3/5/9/6 | 3/8/19/9 | 8/15/19/21 | None | 39 |
| 4 | ATPB, ATP synthase subunitβ, mitochondrial | P10719 | 6/15/8/5 | 7/19/10/5 | 18/45/22/14 | (ATP, GTP) | 56 |
| 5 | EFTU, elongation factor Tu, mitochondrial | P85834 | 4/6/8/3 | 5/8/9/3 | 12/17/21/10 | (GTP, NADH) | 50 |
| 6 | ATAD3, ATPase family AAA domain-containing protein 3 | Q3KRE0 | 6/5/2/2 | 6/5/3/2 | 12/11/4/5 | (ATP) | 67 |
| 7 | ITPR1, inositol 1,4,5-trisphosphate receptor type 1 | P29994 | 2/7/6/10 | 2/7/6/10 | 1.1/3/1.9/6 | (GTP) | 313 |
| 8 | SEPT7, septin-7 | Q9WVC0 | 3/10/3/5 | 3/14/3/7 | 10/27/8/12 | NADH | 51 |
| 9 | GNAO, guanine nucleotide-binding protein Go subunit α | P59215 | 5/7/3/7 | 5/8/3/10 | 18/25/11/25 | (NADH) | 40 |
| 10 | STXB1, syntaxin-binding protein 1 | P61765 | 3/7/2/3 | 3/7/2/3 | 10/16/4/5 | (GTP) | 68 |
| 11 | AP3D1, adaptor-related protein complex 3, δ 1 subunit, isoform CRA_b | B5DFK6 | 3/4/4/6 | 3/5/5/6 | 4/5/5/7 | (NADH) | 136 |
| 12 | AT2B1, plasma membrane calcium-transporting ATPase 1 | P11505 | 3/4/2/7 | 3/4/2/7 | 3/4/2/7 | (NADH) | 139 |
| 13 | AT1A1, sodium/potassium-transporting ATPase subunit α -1 | P06685 | 4/4/1/9 | 4/4/1/12 | 6/5/1.8/11 | (GTP) | 113 |
| 14 | RAB3A, Ras-related protein Rab-3A | P63012 | 4/1/0/2 | 4/1/0/2 | 28/5/0/18 | GTP | 25 |
| 15 | SEPT8, septin-8 | B0BNF1 | 2/2/1/1 | 3/2/1/1 | 6/6/2/4 | NADH | 51 |
| 16 | GBRA1, γ-aminobutyric acid receptor subunit α -1 | P62813 | 2/5/1/0 | 2/5/1/0 | 5/13/2/0 | NADH, (ATP) | 52 |
| 17 | CN37, 2′,3′-cyclic-nucleotide 3′-phosphodiesterase | P13233 | 3/3/1/5 | 3/3/1/5 | 9/9/3/14 | ATP | 47 |
| 18 | SV2A, synaptic vesicle glycoprotein 2A | Q02563 | 2/1/2/3 | 2/1/3/3 | 11/1.5/3/5 | NADH | 83 |
a Nucleotides in parentheses did not give complete competition. Incomplete competition was defined to be the case when two or more assigned spectra but less than the half of those found in capture experiments were identified in the competition experiments.
b MPCP was captured in the presence of any of the three nucleotides used for competition (ATP, GTP, and NADH).
DISCUSSION
Among the different approaches within the field of proteomics research, the isolation of subproteomes based on functional interaction of the target proteins to small molecules, oligonucleotides or peptides, is an increasingly promising approach (30). For this purpose, small molecules may be immobilized on polymer beads (for reviews, see Refs. 31 and 32). Another well established approach is the use of multifunctional activity-based probes that bind selectively and covalently to the active centers of members of the targeted enzyme families. A sorting function, linked to the selectivity group, permits the isolation of the small molecule-protein conjugates. This approach is known as activity-based protein profiling (33, 34). Both techniques have particular strengths and limitations. The affinity bead approach requires relatively large amounts of protein input, preventing its application to scarce samples, such as tissue biopsies or primary cell cultures. In addition, considerable preparative effort is required for achieving sufficient amounts of the input material from appropriate sources. In the activity-based protein profiling approach the small molecule attaches covalently to the target protein to maximize the sensitivity. Therefore, this method is applicable to less abundant protein samples. However, it is restricted to those small molecules that can covalently react with the active centers of targeted proteins, which is not the case for the vast majority of small molecules that specifically bind to target protein families.
We have overcome this restriction by adding a photoactivatable reactivity group to the backbone scaffold attached to the small molecule. A similar approach has been used in distinct cases for the isolation of histone deacetylases from complex protein mixtures (35). We have developed a novel and general approach to functionally isolate and identify proteins by combining the Capture Compound strategy with mass spectrometric analysis and named it CCMS (11). A crucial step in the work flow of our methodology is the photocross-linking under controlled and reproducible conditions, in particular the reaction temperature of the sample as well as wavelength and power of the UV light. For this purpose, we have developed a photocross-linking device with an integrated cooling unit. This allows the photoactivation between 290 and 370 nm (for the spectral characteristics of the UV lamp, see the supplemental material), which is necessary to activate most available reactivity groups beyond the cutoff of cell lysates, at a reaction temperature of 2–4 °C. The lamps emit light with a maximum at 312 nm. Indeed we4 and others (29) found that the well known aryltrifluoromethyldiazirines are most efficiently activated to the carbenes at 310 nm, whereas at 350–360 nm, the wavelength normally used for activation of diazirines, the carbenes are generated only in part alongside transformation of the diazirine to the diazo compound. Here we report the efficiency of CCMS using the novel cAMP-binding protein-specific Capture Compounds as an example. CCMS serves as an analytical tool to probe the cAMP-interacting subset of the proteome. Our results clearly demonstrate that the CCMS strategy can be used to isolate not only principle soluble bona fide cAMP-binding proteins but also their interacting partners, such as the AKAPs.
The sensitivity of the cAMP-CCMS approach was demonstrated by the success in functional isolation and identification of HCN channels from synaptosomal fractions. This result is unique compared to those of existing studies to target cAMP-binding proteins based on small molecule-protein interactions combined with mass spectrometry-based proteomics. It has been reported, based on immunohistochemical data, that HCN1 and HCN2 are localized at the dendritic side of synapses (12, 13). However, to our knowledge, even in large scale comprehensive MS-based screening studies using subcellular proteomics approaches in which more than 2000 individual gene products have been identified from synaptic structures (14–17), HCN channels have escaped analysis. Likewise, in comprehensive cAMP affinity purification experiments from heart ventricular tissue in which HCN4 channels are known to be localized, this class of proteins escaped analysis (8, 9), although other cAMP- and cGMP-binding proteins, such as the regulatory subunits of PKA and the cGMP-dependent protein kinases, were robustly identified in those studies.
To our knowledge there are only a few studies to date that reported the identification of endogenous HCN channels by mass spectrometry-based proteomics at all. One of those is a shotgun-type study to profile phosphopeptides from synaptosome proteins (36) in which a large number of phosphorylation sites of synaptosome proteins were reported, among them a total of seven phosphorylation sites from four different HCN channels. In an entirely different type of experimental approach, Zolles et al. (37) isolated HCN ion channel complexes by an immunoprecipitation-based approach followed by mass spectrometric analysis directed at the identification of new HCN ion channel interaction partners.
This highlights the difficulty to get preparative access to these proteins at the endogenous level in general. Moreover, shotgun approaches as well as immunoprecipitation approaches answer experimental questions different from those addressed by small molecule-protein interaction-based approaches. In contrast to a shotgun approach, the CCMS approach requires still native functional protein during the capture reaction. This is the case in an immunoprecipitation experiment as well, but the latter is entirely targeted to the protein that is recognized by the antibody used. cAMP-CCMS, however, still retains a screening character and addresses a heterogeneous functional subset of the proteome, and sensitivity matters in terms of coverage of the targeted subset.
These observations led us to suggest that CCMS using cAMP-CCs is a uniquely sensitive and efficient strategy to profile target proteins even from samples of relatively low amounts of protein. This further suggests that CCMS using cAMP-CCs permits screening for the target proteins from sample types of restricted abundance, such as tissue biopsies, which cannot be scaled up.
In addition to bona fide cAMP- or cGMP-binding proteins and validated interaction partners (the AKAP proteins), a number of further proteins were captured with 8-AHA-cAMP-CC (7c) but not with scaffold 1, which lacks the selectivity group (Tables III and V). There are three possible explanations for this observation. 1) There may be proteins that specifically bind to cAMP but that lack a CNBD domain. 2) Proteins may have been captured that bind to nucleotides in a promiscuous manner under the experimental conditions chosen. 3) There may be additional indirect binding partners of cAMP/cGMP-binding proteins that were co-isolated through non-covalent interactions. Data from cAMP/cGMP affinity bead pulldown studies or from combined Biacore/LC-MS/MS studies have reported similar results (8, 9, 10). In these studies, the specificity for cyclic nucleotide monophosphate binding has been probed by sequential elution of the bound proteins with a range of different nucleotides, such as ATP, GTP, NAD, and finally cAMP (or cGMP), although elution with cAMP (or cGMP) was often incomplete. In CCMS, the specificity cannot be tested at that stage of the experiment because captured proteins are covalently attached to the Capture Compounds. Instead, in CCMS, the specificity of the Capture Compound-protein interaction can be tested by competition of nucleotides for the binding of the cAMP Capture Compound directly in the CCMS assay. Bona fide cAMP-binding proteins were substantially competed by free 8-AHA-cAMP (4c). Interestingly, ATP at 2 mm was also capable to compete in these cases. This raises the interesting possibility that cAMP signal transduction may be sensitive to the metabolic state of the affected cells and may be “toned down” to a certain extent in situations in which intracellular ATP concentrations are particularly high.
Another major advantage that results from the CCMS work flow is that cAMP-binding proteins can be screened against control samples in which the binding of the CC has truly been competed by a suitable competitor molecule, making the results reliable. This concept is in contrast to the application of cAMP affinity beads. In studies using this affinity bead work flow the specificity of the protein isolation was established through sequential elution of non-cAMP nucleotide-binding proteins by nucleotide compounds such as ADP, NAD, or cGMP. However, the bona fide cAMP-binding proteins could not (8, 9) or could only partially (25) be eluted from the affinity matrix even by cAMP itself. Rather it was concluded that those proteins that were retained on the cAMP bead column even after “elution” with cAMP were the real high affinity cAMP binders. This obviously creates uncertainty in the case where a protein previously not known to interact with cAMP was retained on the affinity beads. CCMS, because of the possibility to efficiently compete for specifically captured proteins, therefore, provides much more solid data in the discovery of novel cAMP-binding proteins.
It may still be possible to scale up the capture experiments by pooling several samples for subsequent protein analysis to increase the number of identified peptides per protein. The scope of the work presented here was to demonstrate the analytical power of CCMS even for low amounts of protein input as well as to identify proteins that are otherwise difficult to access, such as membrane proteins.
Supplementary Material
Acknowledgments
We are in debt to Dr. Daniela Bertinetti and Prof. Friedrich W. Herberg, department of biochemistry, University of Kassel, Kassel, Germany for the determination of EC50 of compound 7c using the fluorescence polarization methodology.
Footnotes
* This work was supported by the European Regional Development Fund of the European Union (Programm für Forschung, Innovation und Technologien ProFIT Grant 10141388).
 The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. F1–F12 and other data.
The on-line version of this article (available at http://www.mcponline.org) contains supplemental Figs. F1–F12 and other data.
2 C. Dalhoff, M. Hüben, T. Lenz, P. Poot, E. Nordhoff, H. Köster, and E. Weinhold, ChemBioChem, accepted.
3 Y. Luo, O. Gràebner (née Baessler), S. Hanke, D. Bertinetti, F. W. Herberg, A. Schrey, M. Glinski, M. Dreger, M. Sefkow, and H. Köster, manuscript in preparation.
4 T. Lenz, unpublished results.
1 The abbreviations used are:
- CNBD
- cyclic nucleotide binding domain
- AKAP
- protein kinase A-anchoring protein
- CAP
- catabolite gene activator protein
- CC
- Capture Compound
- CCMS
- Capture Compound MS
- DIC
- N,N′-diisopropylcarbodiimide
- FA
- formic acid
- HCN
- potassium/sodium hyperpolarization-activated cyclic nucleotide-gated channel
- HepG2
- human hepatocellular liver carcinoma cell line
- HOBt
- N-hydroxybenzotriazole
- PDE
- 3′,5′-cyclic phosphodiesterase
- WB
- wash buffer
- 6-AE-cAMP
- N6-(2-aminoethyl)adenosine 3′,5′-cyclic monophosphate
- 6-AH-cAMP
- N6-(6-aminohexyl)adenosine 3′,5′-cyclic monophosphate
- 2-AEA-cAMP
- 2-(2-aminoethylamino)adenosine 3′,5′-cyclic monophosphate
- 2-AHA-cAMP
- 2-(6-aminohexylamino)adenosine 3′,5′-cyclic monophosphate
- 8-AEA-cAMP
- 8-(2-aminoethylamino)adenosine 3′,5′-cyclic monophosphate
- 8-ABA-cAMP
- 8-(4-aminobutylamino) adenosine 3′,5′-cyclic monophosphate
- 8-AHA-cAMP
- 8-(6-aminohexylamino)adenosine 3′,5′-cyclic monophosphate
- 8-ADOA-cAMP
- 8-(8-amino-3,6-dioxaoctylamino)adenosine 3′,5′-cyclic monophosphate
- PKARII
- protein kinase A, regulatory subunit type II
- PKARIα
- protein kinase A, regulatory subunit type Iα
- LTQ
- linear trap quadrupole
- KAP
- kinase-anchoring protein
- MPCP
- mitochondrial phosphate carrier protein.
REFERENCES
- 1.Beavo J. A., Brunton L. L. ( 2002) Cyclic nucleotide research—still expanding after half a century. Nat. Rev. Mol. Cell Biol 3, 710– 718 [DOI] [PubMed] [Google Scholar]
- 2.McKay D. B., Steitz T. A. ( 1981) Structure of catabolite gene activator protein at 2.9 A resolution suggests binding to left-handed B-DNA. Nature 290, 744– 749 [DOI] [PubMed] [Google Scholar]
- 3.Taylor S. S., Kim C., Cheng C. Y., Brown S. H., Wu J., Kannan N. ( 2008) Signaling through cAMP and cAMP-dependent protein kinase: diverse strategies for drug design. Biochim. Biophys. Acta 1784, 16– 26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hofmann F. ( 2005) The biology of cyclic GMP-dependent protein kinases. J. Biol. Chem 280, 1– 4 [DOI] [PubMed] [Google Scholar]
- 5.Cheng X., Ji Z., Tsalkova T., Mei F. ( 2008) Epac and PKA: a tale of two intracellular cAMP receptors. Acta. Biochim. Biophys. Sin 40, 651– 662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kaupp U. B., Seifert R. ( 2002) Cyclic nucleotide-gated ion channels. Physiol. Rev 82, 769– 824 [DOI] [PubMed] [Google Scholar]
- 7.Wahl-Schott C., Biel M. ( 2009) HCN channels: structure, cellular regulation and physiological function. Cell. Mol. Life Sci 66, 470– 494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Scholten A., Poh M. K., van Veen T. A., van Breukelen B., Vos M. A., Heck A. J. ( 2006) Analysis of the cGMP/cAMP interactome using a chemical proteomics approach in mammalian heart tissue validates sphingosine kinase type 1-interacting protein as a genuine and highly abundant AKAP. J. Proteome Res 5, 1435– 1447 [DOI] [PubMed] [Google Scholar]
- 9.Scholten A., van Veen T. A., Vos M. A., Heck A. J. ( 2007) Diversity of cAMP-dependent protein kinase isoforms and their anchoring proteins in mouse ventricular tissue. J. Proteome Res 6, 1705– 1717 [DOI] [PubMed] [Google Scholar]
- 10.Visser N. F., Scholten A., van den Heuvel R. H., Heck A. J. ( 2007) Semiquantitative identification of cyclic nucleotide-binding proteins from cellular lysates by using a combination of surface plasmon resonance, sequential elution and liquid chromatography-tandem mass spectrometry. ChemBioChem 8, 298– 305 [DOI] [PubMed] [Google Scholar]
- 11.Köster H., Little D. P., Luan P., Muller R., Siddiqi S. M., Marappan S., Yip P. ( 2007) Capture compound mass spectrometry: a technology for the investigation of small molecule protein interactions. Assay Drug Dev. Technol 5, 381– 390 [DOI] [PubMed] [Google Scholar]
- 12.Bender R. A., Kirschstein T., Kretz O., Brewster A. L., Richichi C., Rüschenschmidt C., Shigemoto R., Beck H., Frotscher M., Baram T. Z. ( 2007) Localization of HCN1 channels to presynaptic compartments: novel plasticity that may contribute to hippocampal maturation. J. Neurosci 27, 4697– 4706 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brewster A. L., Chen Y., Bender R. A., Yeh A., Shigemoto R., Baram T. Z. ( 2007) Quantitative analysis and subcellular distribution of mRNA and protein expression of the hyperpolarization-activated cyclic nucleotide-gated channels throughout development in rat hippocampus. Cereb. Cortex 17, 702– 712 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schrimpf S. P., Meskenaite V., Brunner E., Rutishauser D., Walther P., Eng J., Aebersold R., Sonderegger P. ( 2005) Proteomic analysis of synaptosomes using isotope-coded affinity tags and mass spectrometry. Proteomics 5, 2531– 2541 [DOI] [PubMed] [Google Scholar]
- 15.Trinidad J. C., Thalhammer A., Specht C. G., Lynn A. J., Baker P. R., Schoepfer R., Burlingame A. L. ( 2008) Quantitative analysis of synaptic phosphorylation and protein expression. Mol. Cell. Proteomics 7, 684– 696 [DOI] [PubMed] [Google Scholar]
- 16.Li K., Hornshaw M. P., van Minnen J., Smalla K. H., Gundelfinger E. D., Smit A. B. ( 2005) Organelle proteomics of rat synaptic proteins: correlation-profiling by isotope-coded affinity tagging in conjunction with liquid chromatography-tandem mass spectrometry to reveal post-synaptic density specific proteins. J. Proteome Res 4, 725– 733 [DOI] [PubMed] [Google Scholar]
- 17.Li K. W., Hornshaw M. P., Van Der Schors R. C., Watson R., Tate S., Casetta B., Jimenez C. R., Gouwenberg Y., Gundelfinger E. D., Smalla K. H., Smit A. B. ( 2004) Proteomics analysis of rat brain postsynaptic density. Implications of the diverse protein functional groups for the integration of synaptic physiology. J. Biol. Chem 279, 987– 1002 [DOI] [PubMed] [Google Scholar]
- 18.Moll D., Prinz A., Gesellchen F., Drewianka S., Zimmermann B., Herberg F. W. ( 2006) Biomolecular interaction analysis in functional proteomics. J. Neural Transm 113, 1015– 1032 [DOI] [PubMed] [Google Scholar]
- 19.Gray E. G., Whittaker V. P. ( 1962) The isolation of nerve endings from brain: an electron-microscopic study of cell fragments derived by homogenization and centrifugation. J. Anat 96, 79– 88 [PMC free article] [PubMed] [Google Scholar]
- 20.tom Dieck S., Sanmartí-Vila L., Langnaese K., Richter K., Kindler S., Soyke A., Wex H., Smalla K. H., Kämpf U., Fränzer J. T., Stumm M., Garner C. C., Gundelfinger E. D. ( 1998) Bassoon, a novel zinc-finger CAG/glutamine-repeat protein selectively localized at the active zone of presynaptic nerve terminals. J. Cell Biol 142, 499– 509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bradford M. M. ( 1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem 72, 248– 254 [DOI] [PubMed] [Google Scholar]
- 22.Laemmli U. K. ( 1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680– 685 [DOI] [PubMed] [Google Scholar]
- 23.Keller A., Nesvizhskii A. I., Kolker E., Aebersold R. ( 2002) Empirical statistical model to estimate the accuracy of peptide identifications made by MS/MS and database search. Anal. Chem 74, 5383– 5392 [DOI] [PubMed] [Google Scholar]
- 24.Nesvizhskii A. I., Keller A., Kolker E., Aebersold R. ( 2003) A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem 75, 4646– 4658 [DOI] [PubMed] [Google Scholar]
- 25.Bertinetti D., Schweinsberg S., Hanke S. E., Schwede F., Bertinetti O., Drewianka S., Genieser H. G., Herberg F. W. ( 2009) Chemical tools selectively target components of the PKA system. BMC Chem. Biol 9, 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zubay G., Schwartz D., Beckwith J. ( 1970) Mechanism of activation of catabolite-sensitive genes: a positive control system. Proc. Natl. Acad. Sci. U.S.A 66, 104– 110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Popovych N., Tzeng S. R., Tonelli M., Ebright R. H., Kalodimos C. G. ( 2009) Structural basis for cAMP-mediated allosteric control of the catabolite activator protein. Proc. Natl. Acad. Sci. U.S.A 106, 6927– 6932 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Moll D., Zimmermann B., Gesellchen F., Herberg F. W. ( 2006) Current developments for the in vitro characterization of protein interactions, in Proteomics in Drug Research ( Marcus K., Stühler K., van Hall A., Hamacher M., Warscheid B., Meyer H. E. eds) pp. 159– 172, Wiley-VCH Verlag, Weinheim, Germany [Google Scholar]
- 29.Hashimoto M., Hatanaka Y. ( 2006) Practical conditions for photoaffinity labeling with 3-trifluoromethyl-3-phenyldiazirine photophore. Anal. Biochem 348, 154– 156 [DOI] [PubMed] [Google Scholar]
- 30.Hagenstein M. C., Sewald N. ( 2006) Chemical tools for activity-based proteomics. J. Biotechnol 124, 56– 73 [DOI] [PubMed] [Google Scholar]
- 31.Katayama H., Oda Y. ( 2007) Chemical proteomics for drug discovery based on compound-immobilized affinity chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci 855, 21– 27 [DOI] [PubMed] [Google Scholar]
- 32.Bantscheff M., Hopf C., Kruse U., Drewes G. ( 2007) Proteomics-based strategies in kinase drug discovery. Ernst Schering Found. Symp. Proc 1– 28 [DOI] [PubMed] [Google Scholar]
- 33.Fonoviæ M., Bogyo M. ( 2008) Activity-based probes as a tool for functional proteomic analysis of proteases. Expert Rev. Proteomics 5, 721– 730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barglow K. T., Cravatt B. F. ( 2007) Activity-based protein profiling for the functional annotation of enzymes. Nat. Methods 4, 822– 827 [DOI] [PubMed] [Google Scholar]
- 35.Salisbury C. M., Cravatt B. F. ( 2007) Activity-based probes for proteomic profiling of histone deacetylase complexes. Proc. Natl. Acad. Sci. U.S.A 104, 1171– 1176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Munton R. P., Tweedie-Cullen R., Livingstone-Zatchej M., Weinandy F., Waidelich M., Longo D., Gehrig P., Potthast F., Rutishauser D., Gerrits B., Panse C., Schlapbach R., Mansuy I. M. ( 2007) Qualitative and quantitative analyses of protein phosphorylation in naive and stimulated mouse synaptosomal preparations. Mol. Cell. Proteomics 6, 283– 293 [DOI] [PubMed] [Google Scholar]
- 37.Zolles G., Wenzel D., Bildl W., Schulte U., Hofmann A., Müller C. S., Thumfart J. O., Vlachos A., Deller T., Pfeifer A., Fleischmann B. K., Roeper J., Fakler B., Klöcker N. ( 2009) Association with the auxiliary subunit PEX5R/Trip8b controls responsiveness of HCN channels to cAMP and adrenergic stimulation. Neuron 62, 814– 825 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.