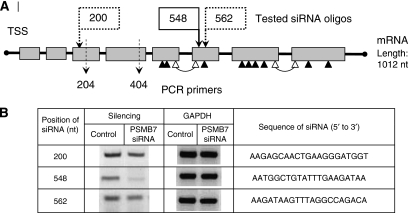
Figure 3.
(A) Schematic view of PSMB7 gene with its eight exons. Splitting positions of tested siRNA oligos are shown (↓); the most effective siRNA is marked by continuously bordered box. Binding location of designed PCR primers specific for PSMB7 is shown under the gene by a broken line (↓). Positions of probe sequences of probeset 200786_at on Affymetrix HG-U133A array are also shown (▴, match probes; ▵, junction probes, overlapping two neighbouring exons). (B) Details of designed siRNA oligos. One out of three siRNA oligos (degrading at 548 nucleotide (nt)) showed effective silencing compared with siRNA-untreated control. GAPDH was used as internal control for both untreated control and siRNA-treated samples. The effect of silencing was 87% for siRNA binding at position 548.