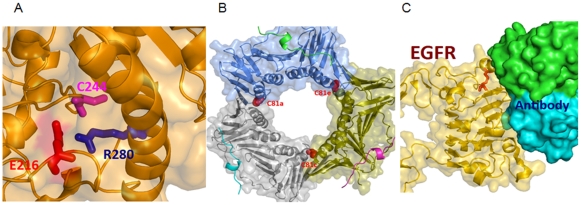
Figure 6. Crystal structure of the representative protein-SNOs.
A) An example of a buried S-nitrosylation site in a crystal structure of the human protein disulfide-isomerase, PDIA3 (PDB code 3F8U), with positively charged (R280) and negatively charged (E216) residues in direct spatial proximity of the nitrosylated cysteine (C244). B) An example of an S-nitrosylation site in direct contact with a ligand and thus likely disrupting/affecting ligand binding upon nitrosylation. One of such examples is EGFR, which is shown here in complex with the monoclonal antibody inhibitor cetuximab (PDB structure 1YY9). Binding of the antibody partially occludes the ligand-binding site in EGFR and keeps it in an inactive conformation. As can be seen from the figure, the targeted cysteine (highlighted using red stick model) directly supports the protein interaction interface in EGFR, suggesting that S-nitrolysation may attenuate its interactions with ligands and inhibitors. C) An example of S-nitrosylation site(s) located within protein-protein (oligomerization) interfaces and that thus may disrupt/attenuate complex formation in PCNA and its function in DNA replication. Note that DNA, which occupies the central hole in the PCNA trimer, is not included in this complex (PDB structure 1VYJ). On the other hand, p21 peptides, which mediate regulatory interactions with CDK/cyclin complexes, are shown using cartoon models.