Abstract
How proteins catalyze morphogenesis is an outstanding question in developmental biology. In bacteria, morphogenesis is intimately linked to remodeling the cell wall exoskeleton. Here, we investigate the mechanisms by which the mother cell engulfs the prospective spore during sporulation in Bacillus subtilis. A membrane-anchored protein complex containing two cell wall hydrolases plays a central role in this morphological process. We demonstrate that one of the proteins (SpoIIP) has both amidase and endopeptidase activities, such that it removes the stem peptides from the cell wall and cleaves the cross-links between them. We further show that the other protein (SpoIID) is the founding member of a new family of lytic transglycosylases that degrades the glycan strands of the peptidoglycan into disaccharide units. Importantly, we show that SpoIID binds the cell wall, but will only cleave the glycan strands after the stem peptides have been removed. Finally, we demonstrate that SpoIID also functions as an enhancer of SpoIIP activity. Thus, this membrane-anchored enzyme complex is endowed with complementary, sequential, and stimulatory activities. These activities provide a mechanism for processive cell wall degradation, supporting a model in which circumferentially distributed degradation machines function as motors pulling the mother cell membranes around the forespore.
Keywords: Morphogenesis, cell shape, peptidoglycan, sporulation, amidase, lytic transglycosylase
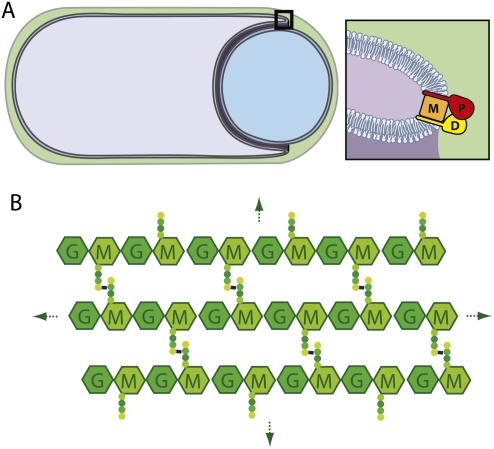
How enzyme complexes orchestrate morphogenic transitions is a fundamental yet poorly understood issue in developmental biology. In eukaryotes, cytoskeletal elements like actin play key roles in controlling changes in cell shape during growth and development (Insall and Machesky 2009). In bacteria, analogous proteins have been implicated in morphogenesis (den Blaauwen et al. 2008). However, since bacteria are encased in an exoskeleton (the cell wall), changes in cell shape ultimately require remodeling of the wall material that surrounds them. The cell wall or peptidoglycan (PG) layer is composed of long glycan chains with repeating units of N-acetylglucosamine (GlcNAc) and N-acetylmuramic acid (MurNAc) linked by glycosidic bonds (Fig. 1B). Attached to the MurNAc sugar is a short peptide that is used to form cross-links between adjacent glycan strands, generating a continuous three-dimensional (3D) meshwork that envelops the bacterium. To remodel this cell-shaped mesh, often referred to as the murein or PG sacculus, enzymes capable of breaking PG linkages are needed. Most bacteria encode a suite of these enzymes, and, in general, every amide and glycosidic linkage in PG is subject to cleavage by at least one family of enzymes that exists in nature (Firczuk and Bochtler 2007; Vollmer et al. 2008).
Figure 1.
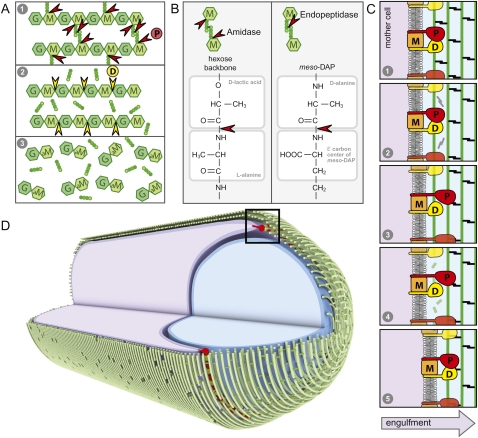
Schematic diagram of the engulfment complex. (A) A sporangium during the morphological processes of engulfment. The membranes of the mother cell (purple) are migrating around the forespore (blue). The cell wall PG encasing the two cells is in green. IID (D), IIP (P), and IIM (M) are all made in the mother cell and localize to the leading edge of the engulfing septal membrane. IID (D) and IIP (P) both have a single N-terminal transmembrane segment and a large extracellular domain. IIM (M) is predicted to have five transmembrane segments. Genetic, biochemical, and cytological analysis indicate that these three proteins reside in a membrane complex (Aung et al. 2007; Chastanet and Losick 2007). (B) Schematic diagram of the PG meshwork. Glycan chains (hexagons) composed of GlcNAc (G) and MurNAc (M) are linked by glycosidic bonds. Attached to the MurNAc (M) sugars are short peptides (balls) that cross-link adjacent glycan strands, generating a continuous 3D meshwork that envelops the bacterium. Gram-positive bacteria like B. subtilis have multiple layers of PG.
The process of sporulation in the bacterium Bacillus subtilis provides a simple and experimentally tractable system in which to study morphogenesis and cell wall remodeling. Upon starvation, B. subtilis differentiates into a dormant cell type called a spore (Stragier and Losick 1996; Errington 2003). The first landmark event in this process is the formation of an asymmetric septum that divides the cell into two unequal-sized compartments: a large cell (called the mother cell) and a small cell (the prospective spore, or forespore). Shortly after polar division, the mother cell membranes migrate around the forespore in a phagocytic-like process, generating a cell within a cell. This morphological process is called engulfment and is the subject of this investigation. Upon the completion of engulfment, the mother nurtures the spore and packages it in a protective coat while the forespore prepares for dormancy. Upon spore maturation, the mother cell lyses, releasing it into the environment.
Three mother cell membrane proteins play a central role in the process of engulfment: SpoIID, SpoIIP, and SpoIIM (hereafter referred to as IID, IIP, and IIM, for simplicity) (Lopez-Diaz et al. 1986; Smith et al. 1993; Frandsen and Stragier 1995). IID and IIP are single-pass transmembrane proteins with large extracellular domains (Fig. 1A). IIM is predicted to have five transmembrane segments and is thought to serve as a scaffold for the assembly of the IID–IIP–IIM complex (Aung et al. 2007; Chastanet and Losick 2007). Cells lacking any one of these proteins are blocked at the earliest stage of the engulfment process: the degradation of the septal PG resident between the mother cell and forespore. Partial loss-of-function mutations in IID or IIP result in defects in the migration of the mother cell membranes around the forespore, supporting the idea that this protein complex is not only involved in removing the septal PG, it also functions in membrane migration (Abanes-De Mello et al. 2002).
Both IID and IIP have been shown to have cell wall-degrading activity in vitro using a gel-based zymography assay (Abanes-De Mello et al. 2002; Chastanet and Losick 2007). However, their enzymatic activities have remained elusive. IIP shares weak similarity to the LytC family of amidases (Pfam_amidase3), and some of the highly conserved residues in IIP are critical for function both in vitro and in vivo (Chastanet and Losick 2007). IID shares sequence similarity with a B. subtilis protein called LytB (Kuroda et al. 1992) that stimulates the amidase activity of LytC (Herbold and Glaser 1975), suggesting that IID could function, in part, to stimulate the proposed amidase activity of IIP (Chastanet and Losick 2007).
All three proteins are synthesized in the mother immediately after polar cell division is complete and localize to the center of the septum, where degradation of the septal PG is initiated (Abanes-De Mello et al. 2002; Aung et al. 2007; Chastanet and Losick 2007). Once the enzyme complexes arrive at the cell periphery, they appear to localize at the leading edge of the engulfing septal membrane (Fig. 1A). It has been postulated that the cell wall degradation activities of these enzyme complexes pull the mother cell membranes (in which these complexes are anchored) around the forespore (Abanes-De Mello et al. 2002). In this model, the PG meshwork serves as a cytoskeletal element directing the enzymatic machinery around the forespore. However, it has remained unclear how the proteins in this complex work together to degrade the septal PG and drive the engulfment process.
Here, we report the biochemical activities of IIP and IID using purified PG in solution-based assays. We demonstrate that IIP has both amidase and endopeptidase activities. Thus, not only does this unusual enzyme remove the stem peptides from PG, it also cleaves the cross-links connecting them. Furthermore, we show that IID is the founding member of a new family of lytic transglycosylases that degrades the glycan strands into disaccharide units. Importantly, our data indicate that IID can bind PG but will only cleave the glycan strands after their stem peptides (and peptide cross-bridges) have been removed. Finally, we show that IID also functions to stimulate IIP activity. Collectively, these activities can be placed into the framework of a catalytic cycle that results in sequential and concerted degradation of the cell wall. Moreover, the observed reaction sequence, along with our finding that both IID and IIP have PG-binding activity, provide a mechanism for processivity of the complex, supporting a model in which circumferentially distributed complexes drive membrane movement around the forespore.
Results
IIP has amidase and endopeptidase activities
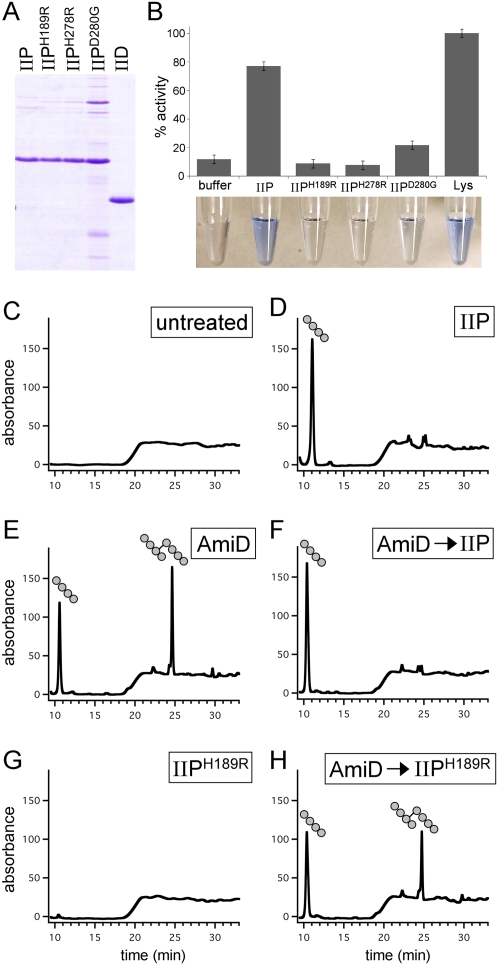
To characterize the PG hydrolase activities of IIP and IID, we used a solution-based dye release assay in which Remazol Brilliant Blue (RBB) was covalently coupled to the sugar moieties of purified Escherichia coli PG. The purified murein sacculi are insoluble and pellet upon centrifugation. However, after cleavage by degradative enzymes, soluble dye-coupled products are generated (Fig. 2B). Absorbance readings of the reaction supernatants after centrifugation serve as a semiquantitative measure of hydrolytic activity (Zhou et al. 1988; T Uehara, T Dinh, and TG Bernhardt, in prep.). The extracellular domain of IIP was fused to a hexahistidine tag and purified from E. coli on Ni2+-agarose (Fig. 2A; see the Materials and Methods). Dye-coupled PG was then incubated with 4 μM purified protein for 30 min at 37°C. Consistent with the reported activity of IIP using a gel-based zymography assay (Chastanet and Losick 2007), we could easily detect the release of soluble PG products (Fig. 2B). Importantly, IIP mutants that have been shown previously to lack activity in vivo and in vitro (Chastanet and Losick 2007) had reduced or undetectable hydrolytic activity in our assay (Fig. 2B). Similar results were obtained using purified sacculi from B. subtilis cells coupled to RBB (Supplemental Fig. S2). However, the release of soluble products by IIP (and other degradative enzymes) using B. subtilis PG was much less efficient and required significantly longer incubation times (data not shown). This could be due to the thicker meshwork of PG in B. subtilis compared with E. coli.
Figure 2.
IIP has both amidase and endopeptidase activity. (A) Coomassie-stained gel of purified proteins. The extracellular domains of wild-type IID, wild-type IIP, and three IIP mutants (Chastanet and Losick 2007) are shown. (B) Cell wall-degrading activities of IIP and IIP mutants using the RBB dye release assay. All reactions contained 4 μM protein. The dye-coupled soluble products released by the IIP proteins for 30 min at 37°C were normalized to the release by 4 μM lysozyme (Lys). Lysozyme cleaves the glycan strands, yielding soluble disaccharide peptide products. Similarly, amidases like AmiD release soluble glycan chains by destroying peptide cross-bridges (Supplemental Fig. S1). The supernatants after incubation with the dye-coupled PG are shown below the histograms. (C–H) RP-HPLC elution profiles of the soluble products after treatment of unlabeled PG with indicated proteins. (C) No treatment of the PG. (D) Incubation with 4 μM IIP. (E) Incubation with 4 μM E. coli AmiD. (F) Incubation with 4 μM AmiD followed by heat inactivation and incubation with 4 μM IIP. Note the loss of cross-linked tetrapeptide after incubation with IIP. (G) Incubation with 4 μM IIPH189R. (H) Incubation with 4 μM AmiD followed by heat inactivation and incubation with 4 μM IIPH189R. Tetrapeptide and cross-linked tetrapeptide products are shown schematically above the elution peaks.
Previous work of Chastanet and Losick (2007) suggested that IIP is an amidase. To directly test this, we incubated the purified protein with unlabeled PG and analyzed the soluble cleavage products by reverse-phase high-performance liquid chromatography (RP-HPLC) (see the Materials and Methods). A single elution peak was detected after treatment with IIP that was not present in the profiles of untreated PG or PG incubated with the IIPH189R mutant (Fig. 2C,D,G). Analysis of the peak fraction at 11 min by mass spectrometry (MS) revealed that the compound had a mass of 462.2 Da, consistent with it being a PG tetrapeptide. These results support the idea that IIP cleaves the amide bond between the lactyl group of muramic acid and the α-amino group of the first amino acid (L-Ala) of the stem peptide. From these experiments, we conclude that IIP is an N-acetylmuramyl-L-alanine amidase (hereafter referred to simply as an amidase).
One striking and unusual feature of the HPLC elution profile of IIP-treated PG was the absence of cross-linked tetrapeptide products. These tetratetrapeptides result from the release of peptide cross-bridges from sacculi and are expected products when amidases act on purified E. coli PG (Fig. 2E). One possible explanation for the absence of this product is that IIP can also act as a D,D-endopeptidase to cleave the peptide cross-link connecting two stem peptides. To investigate this, we first treated the PG with the E. coli amidase AmiD (Uehara and Park 2007) to generate tetrapeptides and cross-linked tetrapeptides (Fig. 2E). After heat inactivation of AmiD, we added IIP to the reaction. Consistent with the idea that IIP possesses endopeptidase activity, the cross-linked tetrapeptide peak (25 min) was lost, and the amount of tetrapeptide product increased (Fig. 2F). Similar endopeptidase activity was observed in experiments in which cross-linked muropeptides generated by a muramidase (Mutanolysin) were used as a substrate for IIP (Supplemental Fig. S3). Importantly, addition of the IIPH189R mutant to the AmiD-treated PG did not result in loss of the tetratetrapeptide product (Fig. 2H). This control indicates that the endopeptidase activity was not due to a contaminating activity in the purified protein. It further shows that His189 is required for both amidase and endopeptidase activities. Altogether, these data indicate that IIP is both an amidase and a D,D-endopeptidase. To our knowledge, this is the first example of an enzyme possessing both activities. We conclude that IIP is the founding member of a new subfamily of the LytC (CwlB) family of amidases (Pfam amidase_3) (Finn et al. 2008). Database searches suggest that this subfamily of amidase/endopeptidase is only present in endospore-forming bacteria.
IID activity requires IIP
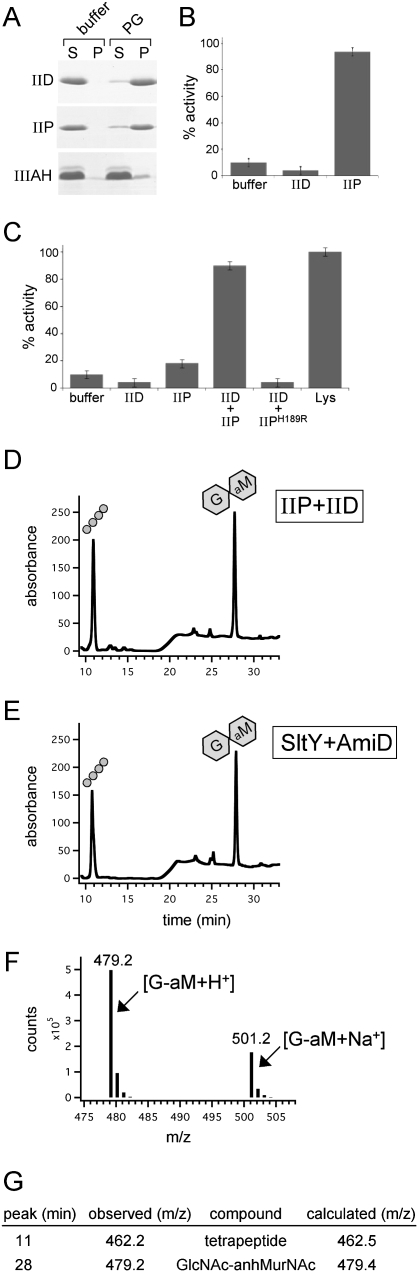
Next, we investigated the PG hydrolase activity of IID. Although the purified extracellular domain of IID (Fig. 2A) efficiently bound PG (Fig. 3A) and produced a zone of clearing in a zymography assay (Abanes-De Mello et al. 2002; data not shown), the protein appeared to have no activity in our solution-based assays (Fig. 3B; Supplemental Fig. S4). Apparently, IID on its own is unable to cleave purified sacculi, or cleavage by IID is not sufficient to generate soluble products. To more rigorously test whether IID cleaves PG, we first incubated the purified protein with unlabeled sacculi, heat-inactivated it, and then treated the PG with well-characterized hydrolytic enzymes that generate defined reaction products with characteristic HPLC elution profiles. If IID cleaves PG, the elution profiles generated from our control enzyme treatments should have new or shifted peaks. On the contrary, the elution profiles after digestion with the E. coli amidase AmiD (Uehara and Park 2007) were indistinguishable, whether or not we first treated the PG with IID (Supplemental Fig. S4). Similar results were obtained using a muramidase (Mutanolysin) that cleaves glycan strands (Supplemental Fig. S4). These experiments suggest that IID alone is unable to cleave purified PG. Based on these findings, we hypothesize that the clear zone observed in the zymography assay with IID is due to IID binding to PG in the gel, resulting in the exclusion of methylene blue from the region where IID migrates, rather than IID generating the zone of clearing by cleaving the immobilized PG. A similar explanation has been proposed recently for E. coli EnvC (T Uehara, T Dinh, and TG Bernhardt, in prep.), which gives a positive zymogram signal but has no detectable PG hydrolase activity in solution-based assays.
Figure 3.
IID activity requires IIP. (A) IID binds PG. The extracellular domains of IID, IIP, and a control protein that does not bind PG (IIIAH) were incubated separately with buffer or E. coli PG. Coomassie-stained gel of the supernatant (S) and pellet (P) fractions after a 16,000g spin is shown. (B) IID has no cell wall-degrading activity on its own. Analysis of IID using the dye release assay. All reactions contained 4 μM protein. The activities of IIP and IID were normalized to lysozyme (data not shown). (C) Analysis of mixtures of IID and IIP using the dye release assay. All reactions with IID and IIP contained 1 μM each protein. The lysozyme reaction contained 4 μM protein. (D) RP-HPLC elution profiles of the soluble products after treatment of unlabeled PG with IIP and IID. The tetrapeptide (balls) and anhydrodisaccharide (hexagons) products are shown schematically above the elution peaks. (E) RP-HPLC elution profiles of the soluble products after treatment of unlabeled PG with SltY and AmiD (see the Materials and Methods). (F) LC/MS analysis in positive ion mode of the product in the 28-min fraction from D. Shown are the mass to charge ratios (m/z) of the ions detected and their relative abundance. (G) Table of calculated and observed m/z of the products in fractions 11 and 28 from D. The product in fraction 11 had an m/z consistent with a tetrapeptide. The product in fraction 28 had an m/z consistent with GlcNAc–anhydro MurNAc.
Since IID and IIP are present in a complex at the site of PG hydrolysis (Abanes-De Mello et al. 2002; Aung et al. 2007; Chastanet and Losick 2007), we wondered whether IID activity requires IIP. To test this, we incubated the two purified proteins with the RBB dye-coupled PG. For these experiments, the concentration of each protein in the reactions was reduced to 1 μM (rather than 4 μM) to prevent complete release of soluble products by IIP during the 30-min incubation. Strikingly, we observed a significant increase in dye release in reactions containing both proteins compared with those with IIP alone (Fig. 3C). Moreover, this activity required catalytically active IIP, since a mixture of the IIPH189R mutant with wild-type IID had no detectable activity (Fig. 3C; Supplemental Fig. S5).
These results suggest either that the PG hydrolytic activity of IID requires IIP or that IID enhances the activity of IIP. To distinguish between these possibilities, we analyzed the soluble products by RP-HPLC. In addition to the elution peak resulting from the amidase activity of IIP, the elution profile contained a new peak (cf. Figs. 2D and 3D). Since IIP has both amidase and endopeptidase activity, we reasoned that this product resulted from cleavage of the glycan strands. Data confirming this idea are presented below. These results are most consistent with a model in which IIP must first remove the peptides from the PG before IID can cleave the glycan strands. However, it is also possible that IIP is required to directly activate IID or that IID activates a latent sugar hydrolase activity in IIP.
IID cleaves glycan strands lacking peptide cross-bridges
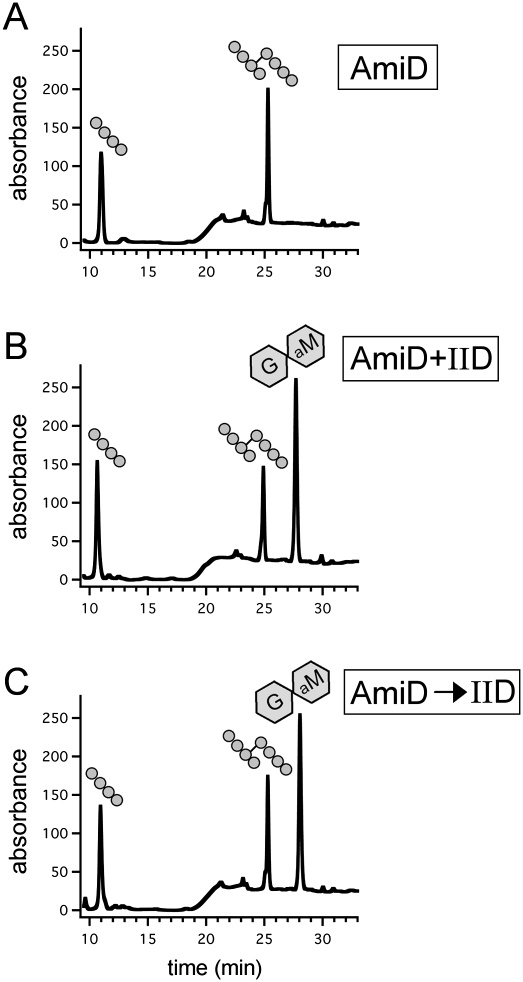
To directly test the model that the amidase activity of IIP generates a substrate for IID, we used an unrelated amidase (AmiD from E. coli) (Uehara and Park 2007) to remove the stem peptides from PG and monitored whether or not IID could cleave the resulting peptide-free or “naked” glycan strands. AmiD is a member of the Pfam amidase_2 family, while IIP is related to the Pfam amidase_3 family; the two proteins share no amino acid sequence similarity. AmiD alone and a mixture of AmiD and IID were incubated with PG, and the reaction products were analyzed by RP-HPLC. As expected, treatment with AmiD generated tetrapeptides and cross-linked tetrapeptides (Fig. 4A). However, in the presence of both IID and AmiD, the elution profile had an additional peak whose elution (at 28 min) was indistinguishable from the new peak observed in the reaction containing IIP and IID (cf. Figs. 3D and 4B). These results support the idea that IID requires denuded glycan strands as a cleavage substrate. To more rigorously test the idea that IID can act on its own to cleave glycan strands lacking stem peptides, we first treated the purified PG with AmiD and then incubated the reaction at 95°C to inactivate the amidase (Supplemental Fig. S6). Next, the naked glycan strands were incubated with IID and the cleavage products were analyzed by RP-HPLC. Consistent with the idea that IID cleaves naked glycan strands, the new cleavage product was efficiently generated (Fig. 4C). Moreover, the amount of cleavage product produced was similar to what was observed when AmiD and IID were incubated simultaneously with PG. Similar results were obtained using IIP (instead of AmiD) in this sequential cleavage assay (Supplemental Fig. S7). Altogether, these results suggest that IID cleaves glycan strands, but only after the stem peptides are removed. This substrate requirement imposes a dependent enzymatic relationship between IIP and IID in the engulfment complex. However, and importantly, the ability of IID to bind the cell wall (Fig. 3A) indicates that IID can bind PG before its cleavage substrate is generated by IIP and can do so independently of IIP.
Figure 4.
IID cleaves naked glycan strands. RP-HPLC elution profiles of the soluble products after treatment of unlabeled PG with the indicated proteins (4 μM each). (A) Incubation with AmiD. (B) Incubation with AmiD and IID. (C) Incubation with AmiD followed by heat inactivation and treatment with IID. The tetrapeptide, cross-linked tetrapeptide, and anhydrodisaccharide products are shown schematically above the elution peaks.
IID is a lytic transglycosylase
There are three known classes of enzymes that cleave glycan strands: muramidases, glucosaminidases, and lytic transglycosylases. Muramidases and glucosaminidases are both sugar hydrolases that cleave the β-1,4 MurNAc–GlcNAc or GlcNAc–MurNAc glycosidic bond, respectively. Lytic transglycosylases, on the other hand, use the C-6 hydroxyl of MurNAc rather than water as a nucleophile in the cleavage reaction of the β-1,4 MurNAc–GlcNAc bond, generating a product ending with 1,6-anhydro MurNAc. IID shares no amino acid sequence similarity to enzymes in any of these three classes. The reaction products of sugar hydrolases and lytic transglycosylases differ by the mass of a water molecule (18 Da). Accordingly, we measured the molecular weight of the IID reaction product to determine whether IID is a hydrolase or a transglycosylase. Using liquid chromatograhpy/MS (LC/MS), the mass to charge ratio (m/z) of the IID reaction product (eluting at 28 min by RP-HPLC) was found to be 479.2 Da, consistent with it being GlcNAc–1,6-anhydro MurNAc (G-aM) (Fig. 3F,G).
To confirm that this product was generated by a lytic transglycosylase, we sought to generate the same anhydrodisaccharide (G-aM) with well-characterized enzymes and compare the retention time of this product by RP-HPLC with the one generated by IID (and IIP). To do this, we first treated the PG with the E. coli lytic transglycosylase SltY (Holtje et al. 1975) to generate G-aM with attached stem peptides. Next, we treated these molecules with the E. coli amidase AmiD to remove the tetrapeptide. The reaction products were then separated by RP-HPLC. In support of the idea that IID is a lytic transglycosylase that cleaves naked glycan strands, the elution profile of SltY/AmiD-treated PG was indistinguishable from PG treated with IID and IIP (Fig. 3, cf. D and E). Altogether, we conclude that IID is the founding member of a new family of lytic transglycosylases.
Amino acid residues required for IID function
Our results showing that IID is a lytic transglycosylase prompted us to search for conserved residues in IID that might be involved in catalysis. Iterative PSI-BLAST searches (Altschul et al. 1997) using B. subtilis IID as the initial query identified a large family of IID homologs (Fig. 5A; Supplemental Fig. S8). Family members are present in both Gram-negative and Gram-positive bacteria, including firmicutes, cyanobacteria, δ-proteobacteria, bacteroidetes, and spirochaetes. In virtually all cases, these proteins have a similar domain structure to B. subtilis IID, containing an N-terminal transmembrane segment followed by a large extracellular domain. The presence of putative IID homologs in non-spore-forming bacteria suggests that, in these organisms, the protein functions in remodeling the PG during vegetative growth.
Figure 5.
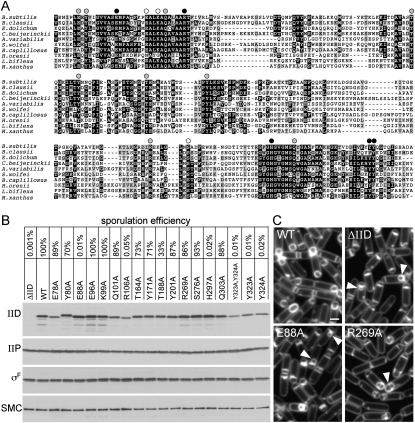
Identification of amino acid residues in IID that are required for function. (A) Amino acid sequence alignment of the extracellular domains of IID homologs from representative phyla. Bacteria include B. subtilis, Bacillus clausii KSM-K16, Eubacterium dolichum DSM 3991, Clostridium beijerinckii NCIMB 8052, Anabaena variabilis ATCC 29413, Syntrophomonas wolfei, Bacteroides capillosus ATCC 29799, Halothermothrix orenii H 168, Leptospira biflexa, and Myxococcus xanthus. See Supplemental Figure S6 for a larger alignment. Conserved amino acids (black boxes) and similar residues (gray boxes) are highlighted. Amino acid substitutions that were tested are indicated above the sequence. Black circles indicate a strong block in engulfment and a >1000-fold defect in sporulation efficiency. Gray circles indicate aberrant engulfment as assessed by fluorescence microscopy. White circles indicate alanine substitutions with no significant affect on engulfment or sporulation efficiency. The first amino acid residue shown for B. subtilis IID is amino acid 72. (B) Sporulation efficiencies of B. subtilis strains containing wild-type IID, a IID-null mutant (ΔIID), and IID mutants with the indicated amino acid substitutions. Cells from the same strains were collected at hour 2 of sporulation, and IID and IIP protein levels were analyzed by immunoblot. All strains efficiently entered sporulation, as judged by the levels of the sporulation transcription factor σF. The SMC protein was used to control for loading. (C) At hour 2 of sporulation, the indicated strains were analyzed by fluorescence microscopy using the membrane dye TMA-DPH. The bulged septal membranes in cells lacking IID or those producing IIDE88A (E88A) are indicated (yellow carets). Impaired engulfment due to the IIDR269A (R269A) mutation is indicated with the white caret. Supplemental Figure S8 shows fluorescence images of all 17 mutants. Bar, 1 μm.
An alignment of the extracellular domains of IID proteins from a diverse group of phyla revealed several highly conserved amino acid residues (Fig. 5A; Supplemental Fig. S8). To determine which, if any, were important for IID function, we introduced alanine substitutions into 17 of the most highly conserved residues that contained polar side chains (Fig. 5A; Supplemental Fig. S8). The IID mutants were introduced in single copy into B. subtilis cells lacking the endogenous IID gene, and the resulting strains were analyzed for their ability to produce heat-resistant spores. Of the 17 mutants tested, five were severely impaired in sporulation efficiency (Fig. 5B). These included alanine substitutions at Glu 88 (E88A), Arg 106 (R106A), His 297 (H297A), and Tyr 323 and Tyr 324 (Y323A and Y324A). In addition to these five mutants, several others had modest defects in sporulation efficiency (Fig. 5B). Importantly, most of the mutants produced IID protein at levels indistinguishable from wild type (Fig. 5B). Although some of the mutants had reduced protein levels (approximately threefold to fourfold lower than wild type), these levels were sufficient in at least one mutant (E78A) to support efficient sporulation (Fig. 5B). In addition, the levels of IID's enzymatic partner IIP were unaffected by the mutations in IID (Fig. 5B).
To determine the stage at which the IID mutants were blocked in the sporulation pathway, we analyzed them by fluorescence microscopy. Cells were induced to sporulate by resuspension in defined minimal media and were analyzed in a sporulation time course. The five mutants with strong defects in sporulation efficiency all phenocopied the IID-null mutant (Fig. 5C; Supplemental Fig. S9). At hour 2 of sporulation, most wild-type cells have initiated engulfment, and many have completed this morphological process (Fig. 5C). In contrast, at this time point, all five mutants had flat septa (indicative of a failure to degrade the septal PG) or contained a characteristic membrane bulge (see yellow carets in Fig. 5C). The bulge phenotype is thought to result from a small amount of degradation of PG at the center of the septum. As sporulation continued, these phenotypes persisted and eventually led to forespore collapse (data not shown). Cytological analysis of sporulating cells can reveal subtle defects that are missed by analyzing spore heat resistance, a terminal phenotype. Accordingly, we analyzed the remaining 12 mutants that had relatively modest or undetectable sporulation defects. Interestingly, nine of these mutants were partially impaired in the engulfment process (Fig. 5; Supplemental Fig. S9). These mutants exhibited a delay in the dissolution of the septal PG as well as defects in migration of the membranes around the forespore (see white caret in Fig. 5C; Supplemental Fig. S9). As originally put forward by Pogliano and coworkers (Abanes-De Mello et al. 2002), these phenotypes support the idea that IID functions not only in removal of septal PG, but also in the engulfment process itself. Altogether, these results indicate that several highly conserved amino acids are important for the full function of IID. Moreover, the data show that residues E88, H297, R106, Y323, and Y324 are essential for IID activity in vivo, and suggest that these residues might serve as catalytic residues or reside at the catalytic center of the lytic transglycosylase.
IID stimulates IIP activity
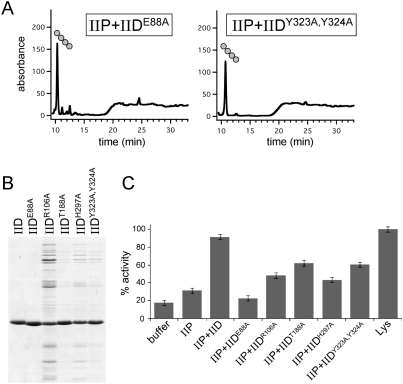
To investigate whether any of the IID mutants that impair function in vivo were defective in cleaving glycan strands, we purified the extracellular domains of several IID mutants (Fig. 6B) and analyzed their activities in vitro (Fig. 6A). Since IID cannot cleave glycan strands that have stem peptides, these assays were carried out in the presence of wild-type IIP. All mutants tested produced little or no anhydrodisaccharide products, as determined by RP-HPLC (Fig. 6A; data not shown), supporting the idea that the residues that are required for IID function in vivo are important for lytic transglycosylase activity.
Figure 6.
IID enhances IIP activity. (A) RP-HPLC elution profiles of the soluble products after treatment of unlabeled PG with IIP and the indicated IID mutants. The tetrapeptide product is shown schematically. (B) Coomassie-stained gel of purified proteins. The extracellular domains of wild-type IID and five IID mutants are shown. (C) Analysis of IIP activity in the presence of the IID mutants using the RBB dye release assay. All reactions with IID and IIP contained 1 μM each protein. The lysozyme reaction contained 4 μM protein.
IID shares weak similarity with B. subtilis LytB (Kuroda et al. 1992), a protein that has been shown to enhance the amidase activity of B. subtilis LytC (Herbold and Glaser 1975). Based on this sequence similarity, IID homologs in other bacteria have been annotated as amidase enhancers (or sometimes misannotated as amidases). Interestingly, virtually none of the amino acid residues that are highly conserved among IID family members are present in LytB. Moreover, none of the amino acids that are critical for the lytic transglycosylase activity of IID are conserved in LytB (Supplemental Fig. S10), suggesting that LytB does not possess this activity. Consistent with this idea, earlier studies failed to detect cell wall-degrading activity for this protein (Herbold and Glaser 1975). However, because of the similarity between LytB and IID, we wondered whether IID might enhance the activity of its cognate amidase IIP. To investigate this, we took advantage of the IID mutants that lack lytic transglycosylase activity. Wild-type IIP was mixed with the IID mutants and incubated with the RBB dye-coupled PG. Interestingly, some of the mutants were able to enhance IIP activity, resulting in up to 2.5-fold greater dye release (Fig. 6C). This degree of stimulation is similar to the magnitude of LytB enhancement of LytC (Herbold and Glaser 1975). These results indicate that IID is not only a lytic transglycosylase that cleaves naked glycan strands, it is also an enhancer of the amidase that generates its substrate.
Discussion
Here, we defined the biochemical activities of the cell wall-degrading enzymes in the engulfment complex. We showed that IIP is an unusual PG hydrolase that possesses both amidase and endopeptidase activities. We further demonstrated that IID is a novel lytic transglycosylase. Thus, this complex is endowed with complementary enzymatic activities to fully disassemble the PG meshwork (Fig. 7A). Interestingly, and importantly, IID (like IIP) can bind the cell wall, but will not cleave glycan strands until the stem peptides have been removed. This requirement for denuded glycan strands enforces an ordered cleavage reaction within the engulfment complex: first IIP, then IID. Finally, our data indicate that IID also functions to stimulate IIP activity. Collectively, these activities can be placed into the framework of a catalytic cycle (described below) resulting in the sequential and concerted degradation of the cell wall. Thus, the engulfment complex represents the first example of a coordinated cell wall-degrading enzyme complex. Furthermore, the observed reaction sequence, along with our finding that both factors have PG-binding activity, provides a mechanism for processivity of the complex, and therefore supports a model (Abanes-De Mello et al. 2002) in which circumferentially distributed complexes drive membrane movement around the forespore.
Figure 7.
Coordinated activities in the engulfment complex drive membrane migration around the forespore. (A) Schematic representation of the complementary and sequential activities of IIP and IID. (1) In the diagram, IIP cleaves the stem peptide from the glycan strand and the cross-links between the tetrapeptides. (2) Following peptide cleavage by IIP, IID cleaves the denuded glycan strands into anhydrodisaccharides. (B) Comparison of the amide bonds cleaved by IIP. (C) Schematic diagram of the proposed catalytic cycle of the engulfment complex. (1) IIP (P) and IID (D) in complex with IIM (M) bind the PG. (2) IID stimulates the amidase activity of IIP, resulting in cleavage of the stem peptides and peptide cross-links (black). (3) IIP is released from the PG and rebinds at a nearby site. (4) IID cleaves the denuded glycan strands (green). (5) IID is released from the PG and rebinds adjacent to IIP. The leading edge of the engulfing mother cell membrane (dark purple) moves toward the forespore pole (to the right). (D) Circumferentially distributed engulfment complexes drive membrane movement around the forespore. Shown is a schematic diagram of a sporangium during the morphological processes of engulfment. The membranes of the mother cell (purple) are migrating around the forespore (blue). The glycan strands (green) are shown running perpendicular to the long axis of the cell, as has been proposed for E. coli and C. crescentus (Holtje 1998; Gan et al. 2008). The hoops of glycan strands are stitched together by the cross-linked tetrapeptides. IID (yellow) and IIP (red) are anchored in the leading edge of the mother cell membrane. Processive degradation of the PG drives the mother cell membranes toward the cell pole. For simplicity, we show the engulfment complex acting on the basement layer of the PG meshwork. It is possible that the complex degrades more than one layer, and not necessarily the layer adjacent to the forespore.
IIP is both an amidase and an endopeptidase
IIP shares weak similarity with CwlV and PlyPSA, two members of the LytC (CwlB) family of amidases (Pfam amidase_3) (Chastanet and Losick 2007; Finn et al. 2008). However, based on the limited sequence similarity, IIP appears to represent a new subfamily of this group. Our biochemical analysis confirms that IIP has amidase activity. Interestingly, IIP is also able to cleave the amide bond that cross-links the stem peptides from adjacent glycan strands, an activity thought to be the purview of dedicated D,D-endopeptidases. Moreover, the same histidine residue that is thought to be part of the catalytic center of the enzyme is required for both activities, suggesting that IIP employs a single catalytic center. Although initially surprising, closer examination of the cleavage substrates reveals a similar chemical structure in the neighborhood of the scissile bonds (Fig. 7B). Thus, the same active site could indeed carry out both hydrolytic reactions. It remains unknown why IIP possesses endopeptidase activity and what role this cleavage plays in sporulation. However, one possibility may be that the endopeptidase activity helps starving sporangia recycle or metabolize the cross-linked peptides so that this source of amino acids is not wasted.
IID is the founding member of a new family of lytic transglycosylases
Initially, we were able to detect only PG-binding activity for IID, not PG cleavage (Fig. 3A). However, upon adding IIP or the unrelated E. coli amidase AmiD to the PG hydrolysis reactions, we discovered that IID is a lytic transglycosylase that requires peptide-free or “naked” glycan chains as a substrate (Figs. 3, 4). Since IID shares no sequence similarity with any PG-degrading enzymes, our work establishes it as the founding member of a new family of lytic transglycosylases. IID family members are found in Gram-negative and Gram-positive bacteria. In most cases, these bacteria are not spore formers, suggesting that these homologs function during vegetative growth to remodel the cell wall. In addition to the IID family, four other families of lytic transglycosylases have been identified (Blackburn and Clarke 2001). Structural studies of representatives of each family have revealed that members of families 1, 3, and 4 (SltY, MltB, and λR, respectively) are all related to goose egg white lysozyme based on their active site folds. All three have catalytic glutamate residues (Vollmer et al. 2008). The structure of MltA, a member of family 2, revealed that this family has a unique fold and a catalytic aspartate residue (van Straaten et al. 2007). We identified Glu 88 as essential for IID activity in vivo and in vitro (Figs. 5, 6), suggesting that this residue serves as the catalytic residue for this enzyme. Compared with other lytic transglycosylases, IID appears to be unique in its requirement for denuded glycan strands as substrates and in its ability to activate the production of its substrate by another enzyme, IIP.
One consequence of the substrate requirement of IID is that muropeptides (disaccharides linked to stem peptides) are not generated by the engulfment complex. This is significant because it has been shown recently that muropeptides can trigger germination of B. subtilis spores through an interaction with a receptor located in the inner forespore membrane (Shah et al. 2008). The sequential activities of IIP and IID would therefore ensure that muropeptide germinants would not be produced during engulfment, thus preventing premature and inappropriate germination.
There is one apparent inconsistency between previous genetic analysis and our biochemical characterization of IID and IIP. Cells lacking either of these two proteins have indistinguishable phenotypes (Eichenberger et al. 2001; Abanes-De Mello et al. 2002). Shortly after polar division, both are blocked in dissolution of the septal PG. Moreover, at later time points, both mutants generate membrane bulges, presumably due to degradation of a small amount of PG at the center of the septum that allows the forespore membranes to invade the mother cell compartment. The prevailing view is that, in the absence of one of these enzymes, the other has a basal level of activity that accounts for the septal bulging. In support of this idea, cells lacking both IID and IIP have flat septa that bulge infrequently (Eichenberger et al. 2001). In contrast, our data showing that IID requires denuded glycan strands to initiate cell wall degradation while IIP has no apparent requirements predict that cells lacking IIP should have a stronger block to engulfment and fewer septal bulges than cells lacking IID. One explanation for the similar mutant phenotypes is that, in the absence of IID, IIP activity is reduced, as we saw in vitro (Fig. 6C). In addition, it is possible that, in the absence of IIP, other amidases present in the septal membrane generate small stretches of denuded glycan strands that IID can cleave to generate PG lesions and the resulting membrane bulges.
A coordinated enzymatic complex drives engulfment around the forespore
Our data indicate that IID and IIP have complementary, sequential, and stimulatory activities. Moreover, both proteins have PG-binding activity (Fig. 3A). We propose that these activities constitute a catalytic cycle for the engulfment complex that is shown schematically in Figure 7C and has the following reaction sequence: (1) IID and IIP (in complex with IIM) bind the cell wall; (2) IID stimulates IIP-dependent peptide cleavage; (3) IIP is released from the PG layer and reassociates with the next available binding site while IID remains bound; (4) release of IIP exposes the naked glycan strand, allowing IID to cleave it; (5) IID is, in turn, released from the PG and rebinds adjacent to IIP. Importantly, at any given time during this catalytic cycle, at least one member of the engulfment complex remains bound to the PG layer such that degradation proceeds processively. Repetition of this cycle by circumferentially distributed complexes (Fig. 7D) would, thus, progressively step the leading edge of the mother cell membrane around the forespore compartment. In this model, the movement of the complex along the PG meshwork could be brought about by conformational changes in IIP and/or IID that allow the proteins to “reach” or extend to capture the next binding site. Alternatively, small changes in the position of the leading edge of the septal membrane brought about by thermal motion could be sufficient.
For the purposes of our schematics, we depicted the glycan strands running perpendicular to the long axis of the cell, as has been proposed for E. coli and Caulobacter crescentus (Holtje 1998; Gan et al. 2008). However, our model is not dependent on this configuration, and works equally well if the glycan strands are oriented along the long axis (Abanes-De Mello et al. 2002). In this case, an engulfment complex would move processively along a single glycan strand, first removing the peptide cross-links (IIP) and then cleaving the denuded glycan chain (IID), leaving disaccharide units and tetrapeptides in its wake.
One outstanding question that is not addressed by our data is: What dictates the direction of the engulfment complex? (Why does it move toward the pole and around the forespore, rather than toward midcell?) One possibility is that a protein in the forespore membrane stimulates the cell wall-degrading activities of the engulfment complex drawing it toward the cell pole. Alternatively, it has been hypothesized that the cell wall has an inherent orientation, and this orientation may be responsible for directing the enzymatic machinery toward the pole (Abanes-De Mello et al. 2002).
The use of the cell wall as a cytoskeletal element to direct membrane movement through the action of cell wall hydrolases was originally proposed by Pogliano and coworkers (Abanes-De Mello et al. 2002). Here, we provided a mechanism for the coordinated movement of the engulfment complex around the forespore. We hypothesize that membrane-anchored cell wall-degrading machines could be employed more generally to move membranes. Specifically, we propose that a similar strategy is used by Gram-negative bacteria to help drive constriction of the outer membrane during cell division. In this case, PG hydrolases tethered to the outer membrane would promote the inward movement of the outer membrane as they split the septal PG layer.
Materials and methods
General methods
All B. subtilis strains were derived from the prototrophic strain PY79 (Youngman et al. 1983). Sporulation was induced by resuspension at 37°C according to the method of Sterlini-Mandelstam (Harwood and Cutting 1990) or by exhaustion (in supplemented DS medium) (Schaeffer et al. 1965). Sporulation efficiency was determined in 36-h cultures as the total number of heat-resistant (20 min at 80°C) colony-forming units (CFUs) compared with wild-type heat-resistant CFUs. Whole-cell lysates from sporulating cells (induced by resuspension) were prepared as described previously (Doan et al. 2009). Samples were heated for 10 min at 50°C prior to loading. Equivalent loading was based on OD600 at the time of harvest. Tables of strains, plasmids, and oligonucleotide primers and descriptions of plasmid construction can be found in the Supplemental Material.
Protein purification
All fusion proteins were expressed in E. coli BL21(DE3). All protein purification steps were carried out at 4°C. For overproduction of the extracellular domain of SpoIID (and SpoIID mutants), cells were grown in LB supplemented with ampicillin (50 μg/mL) at 37°C to an OD600 of 0.6 and induced by the addition of IPTG to 0.5 mM. The cells were harvested after 2 h. For overproduction of the extracellular domain of IIP (and IIP mutants), cells were grown at 16°C to an OD600 of 0.4 and induced for 16 h. Cells were harvested by centrifugation and resuspended in 1/40th vol of buffer Q (20 mM Tris-HCl at pH 8, 300 mM NaCl, 20 mM imidazole, 5 mM 2-mercaptoethanol, 1 mM PMSF), and were lysed by two passages through a French pressure cell. A soluble fraction was prepared by a 100,000g spin and was loaded on a 2-mL Ni2+ resin spin column (NUNC ProPur) equilibrated with buffer Q. Bound protein was washed with buffer W (20 mM Tris-HCl at pH 8, 300 mM NaCl, 20 mM imidazole, 10% glycerol, 5 mM 2-mercaptoethanol, 1 mM PMSF) and eluted in buffer W containing 300 mM imidazole. Peak fractions were pooled and concentrated, and the buffer was exchanged using Millipore filter units (10-kDa cutoff) into 20 mM Tris-HCl (pH 8), 300 mM NaCl, and 10% glycerol. Protein concentration was determined by Bradford and SDS-PAGE followed by Coomassie staining. Purified IIP was used as an antigen to generate anti-IIP rabbit polyclonal antibodies (Covance). Crude serum was affinity-purified as described previously (Campo and Rudner 2006).
For purification of AmiD, cells were grown in LB supplemented with kanamycin (20 μg/mL) and glucose (0.04%) at 37°C to an OD600 of 1.1. IPTG was then added to 1 mM, and growth was continued for an additional 4 h. The cells were harvested by centrifugation and resuspended in buffer A (50 mM Tris-HCl at pH 8.0, 300 mM NaCl, 10% glycerol) with 20 mM imidazole. After a freeze–thaw cycle, the cells were lysed by two passages through a French pressure cell. A soluble fraction was prepared by centrifugation with 100,000g, and the cell extract was loaded onto a NiNTA FF column (GE Healthcare) pre-equilibrated with buffer A with 20 mM imidazole using an AKTA purifier 10. The column was washed with buffer A with 20 mM imidazole, and the protein was eluted with a linear 20 mM to 0.3 M gradient of imidazole in buffer A over 30 column volumes. Peak fractions were collected, desalted with a Hiprep 26/10 column (GE Healthcare), and eluted in buffer B (20 mM Tris-HCl at pH 8.0, 10% glycerol). The peak fraction was collected and loaded onto a Resource Q column (GE Healthcare). The column was washed with buffer B, and the protein was eluted with a linear 0–0.5 M gradient of NaCl in buffer B over 10 column volumes. The peak fraction was dialyzed against 20 mM Tris-HCl (pH 8.0), 100 mM NaCl, and 50% glycerol.
SltY was purified using a His-SUMO purification system as described previously (Bendezu et al. 2009). Cells were grown in LB supplemented with ampicillin (50 μg/mL) and glucose (0.04%) at 37°C to an OD600 of 1.1, IPTG was added to 0.5 mM, and growth was continued for an additional 3 h. Cells were harvested, resuspended in buffer A with 20 mM imidazole, and lysed using a French pressure cell. The cell extract was obtained by centrifugation with 100,000g and loaded onto a 2-mL Ni2+ resin spin column (NUNC ProPur). The column was washed with buffer A with 50 mM imidazole, and the protein was eluted with buffer A with 300 mM imidazole. During overnight dialysis of the eluted protein against buffer A, the protein was cleaved by incubation with His6-tagged SUMO protease (Bendezu et al. 2009). The cleaved protein solution was loaded onto a Ni2+ resin (NUNC ProPur) equilibrated with buffer A. The flow-through fraction containing untagged SltY was collected and concentrated with a Spin-X UF concentrator (10-kDa cutoff; Corning). The protein concentrations of AmiD and SltY were measured with NI protein assay (G-Bioscience).
Immunoblot analysis
Proteins were separated by SDS-PAGE on 12.5% polyacrylamide gels, electroblotted onto Immobilon-P membranes (Millipore), and blocked in 5% nonfat milk in phosphate-buffered saline (PBS)–0.5% Tween-20. The blocked membranes were probed with anti-IID (1:10,000) (Doan and Rudner 2007), anti-IIP (1:5000), anti-σF (1:5000) (Pan et al. 2001), and anti-SMC (1:5000) (Lindow et al. 2002) diluted into 3% BSA in 1× PBS–0.05% Tween-20. Primary antibodies were detected using horseradish peroxidase-conjugated goat, anti-rabbit IgG (Bio-Rad) and the Western Lightning reagent kit as described by the manufacturer (PerkinElmer).
Fluorescence microscopy
Fluorescence microscopy was performed with an Olympus BX61 microscope as described previously (Doan et al. 2009). Fluorescent signals were visualized with a phase contrast objective UplanF1 100× and captured with a monochrome CoolSnapHQ digital camera (Photometrics) using Metamorph software version 6.1 (Universal Imaging). The membrane dye TMA-DPH (Molecular Probes) was used at a final concentration of 0.01 mM, and exposure times were typically 200 msec. Images were analyzed, adjusted, and cropped using Metamorph software.
Preparation of E. coli sacculi labeled with RBB
Sacculi were prepared from E. coli strain TU163 [Δlpp] as described previously (T Uehara, T Dinh, and TG Bernhardt, in prep.). Briefly, 1 L of cells grown in LB was harvested at an OD600 of 0.5 and boiled in 100 mL of 4% SDS with vigorous stirring for 30 min. Sacculi were then harvested by ultracentrifugation and washed five times with ddH2O to remove SDS. Isolated sacculi were treated with 200 μg/mL amylase (Sigma) overnight at 37°C in 1× PBS. The amylase-treated sacculi were washed with ddH2O and then incubated with 20 mM RBB (Sigma) in 0.25 M NaOH overnight at 37°C. The preparation was neutralized with HCl, and the dye-labeled sacculi were pelleted by centrifugation (21,000g, 20 min, room temperature). The sacculi were then washed repeatedly with ddH2O until the supernatant was clear. The final pellet was resuspended in 1 mL of ddH2O containing 0.02% azide and stored at 4°C.
Preparation of B. subtilis sacculi labeled with RBB
Sacculi from a B. subtilis strain derived from the prototrophic strain PY79 were prepared as described previously (McPherson and Popham 2003). Briefly, 200 mL of cells grown in LB were harvested at an OD600 of 0.5 and boiled in 100 mL of 4% SDS with vigorous stirring for 30 min. Sacculi were then harvested by centrifugation (8000g, 6 min, room temperature) and washed eight times with ddH2O to remove SDS. Isolated sacculi were treated with 10 μg/mL DNase I (Worthington Biochemical) and 50 μg/mL RNase A (USB Corporation) for 2 h at 37°C in buffer T (100 mM Tris HCl at pH 7.5, 20 mM MgSO4), and with 100 μg/mL trypsin (Worthington Biochemical) overnight at 37°C in buffer T containing 10 mM CaCl2. The sacculi were boiled again in 1% SDS for 15 min, diluted in 7 mL of ddH2O, and centrifuged at 12,000g for 10 min at room temperature. The pelleted sacculi were then washed twice in 20 mL of ddH2O, once in 20 mL of 8 M LiCl, and twice again in 20 mL of ddH2O. The final pellet was resuspended in 2 mL of water containing 0.02% azide and stored at 4°C. The B. subtilis sacculi were labeled with RBB using the same protocol as was described for E. coli sacculi.
Dye release assay
Ten microliters to 20 μL of RBB-labeled sacculi were incubated for 30 min at 37°C with purified proteins in 100 μL of 1× PBS. Final protein concentrations were 1–4 μM and are indicated in the legends for each figure. Reactions were terminated by incubation for 5 min at 95°C. Soluble cleavage products were separated by centrifugation (21,000g for 20 min) at room temperature. The supernatant was collected, and the absorbance was measured at 595 nm.
PG cleavage assay for RP-HPLC analysis.
Wild-type sacculi from E. coli strain TB28 were prepared as described previously (Uehara et al. 2009). Unless otherwise noted, the reaction was performed overnight at 37°C in 50 μL of 10 mM HEPES-NaOH (pH 7.2) containing 10 μL of sacculi and 4 μM purified protein. The reactions were stopped by the addition of 20 μL of 20% phosphoric acid. When reduction of samples was carried out, the reaction was incubated with ∼1 mg of sodium borohydride in 50 mM sodium borate (pH 9.0) for 30 min at room temperature, followed by addition of 20% phosphoric acid to adjust pH to <4. For sequential digestion experiments, sacculi were treated overnight at 37°C with the first enzyme. After heat inactivation for 15 min at 95°C, the sample was incubated overnight at 37°C with the second enzyme. Sequential digestion of sacculi with SltY and AmiD was performed with 6 μM SltY in 20 mM potassium-acetate buffer (pH 4.5). After heat inactivation, 20 mM HEPES-NaOH (pH 7.2) and 4 μM AmiD were added for the second digestion reaction. All of the samples were centrifuged (21,000g, 10 min) prior to RP-HPLC analysis.
RP-HPLC analysis of products from PG cleavage reactions
Analyses of the reduced and nonreduced samples by RP-HPLC were performed with a Dionex Ultimate 3000 system equipped with a Jupiter Proteo C12 column (250 mm × 4.6 mm, 4 μm, 90 Å; Phenomenex) as described previously (Uehara et al. 2009). To identify the products that eluted at 11 min and 28 min in Figure 3D, the fraction was analyzed with a 6520 Accurate-Mass quadrupole time-of-flight LC/MS (Agilent) with a Jupiter Proteo C12 column (250 mm × 4.6 mm, 4 μm, 90 Å; Phenomenex). The samples were injected onto the column and eluted at a flow rate of 0.5 mL/min with 1% formic acid in water for 5 min, followed by a linear gradient of 1%–10% methanol over 10 min.
Acknowledgments
We thank members of the Rudner and Bernhardt laboratories past and present for valuable discussions and support. We acknowledge Emma Doud and Suzanne Walker for assistance with LC/MS, Janet Iwasa for help with digital art, and Matt Waldor and Mike Springer for valuable discussions. We thank Enki Morlot Boyault for waiting until almost all the data were generated to be born. Support for this work comes in part from the National Institutes of Health Grant GM073831-01A1 (to D.Z.R.); Burroughs Welcome Fund, Massachusetts Life Science Center, and National Institutes of Health Grant AI08365-01A1 (to T.G.B.); and the Giovanni Armenise-Harvard Foundation (to D.Z.R. and T.G.B.).
Footnotes
Article is online at http://www.genesdev.org/cgi/doi/10.1101/gad.1878110.
Supplemental material is available at http://www.genesdev.org.
References
- Abanes-De Mello A, Sun YL, Aung S, Pogliano K. A cytoskeleton-like role for the bacterial cell wall during engulfment of the Bacillus subtilis forespore. Genes & Dev. 2002;16:3253–3264. doi: 10.1101/gad.1039902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aung S, Shum J, Abanes-De Mello A, Broder DH, Fredlund-Gutierrez J, Chiba S, Pogliano K. Dual localization pathways for the engulfment proteins during Bacillus subtilis sporulation. Mol Microbiol. 2007;65:1534–1546. doi: 10.1111/j.1365-2958.2007.05887.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendezu FO, Hale CA, Bernhardt TG, de Boer PA. RodZ (YfgA) is required for proper assembly of the MreB actin cytoskeleton and cell shape in E. coli. EMBO J. 2009;28:193–204. doi: 10.1038/emboj.2008.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn NT, Clarke AJ. Identification of four families of peptidoglycan lytic transglycosylases. J Mol Evol. 2001;52:78–84. doi: 10.1007/s002390010136. [DOI] [PubMed] [Google Scholar]
- Campo N, Rudner DZ. A branched pathway governing the activation of a developmental transcription factor by regulated intramembrane proteolysis. Mol Cell. 2006;23:25–35. doi: 10.1016/j.molcel.2006.05.019. [DOI] [PubMed] [Google Scholar]
- Chastanet A, Losick R. Engulfment during sporulation in Bacillus subtilis is governed by a multi-protein complex containing tandemly acting autolysins. Mol Microbiol. 2007;64:139–152. doi: 10.1111/j.1365-2958.2007.05652.x. [DOI] [PubMed] [Google Scholar]
- den Blaauwen T, de Pedro MA, Nguyen-Disteche M, Ayala JA. Morphogenesis of rod-shaped sacculi. FEMS Microbiol Rev. 2008;32:321–344. doi: 10.1111/j.1574-6976.2007.00090.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Rudner DZ. Perturbations to engulfment trigger a degradative response that prevents cell-–cell signalling during sporulation in Bacillus subtilis. Mol Microbiol. 2007;64:500–511. doi: 10.1111/j.1365-2958.2007.05677.x. [DOI] [PubMed] [Google Scholar]
- Doan T, Morlot C, Meisner J, Serrano M, Henriques AO, Moran CP, Jr, Rudner DZ. Novel secretion apparatus maintains spore integrity and developmental gene expression in Bacillus subtilis. PLoS Genet. 2009;5:e1000566. doi: 10.1371/journal.pgen.1000566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichenberger P, Fawcett P, Losick R. A three-protein inhibitor of polar septation during sporulation in Bacillus subtilis. Mol Microbiol. 2001;42:1147–1162. doi: 10.1046/j.1365-2958.2001.02660.x. [DOI] [PubMed] [Google Scholar]
- Errington J. Regulation of endospore formation in Bacillus subtilis. Nat Rev Microbiol. 2003;1:117–126. doi: 10.1038/nrmicro750. [DOI] [PubMed] [Google Scholar]
- Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firczuk M, Bochtler M. Folds and activities of peptidoglycan amidases. FEMS Microbiol Rev. 2007;31:676–691. doi: 10.1111/j.1574-6976.2007.00084.x. [DOI] [PubMed] [Google Scholar]
- Frandsen N, Stragier P. Identification and characterization of the Bacillus subtilis spoIIP locus. J Bacteriol. 1995;177:716–722. doi: 10.1128/jb.177.3.716-722.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan L, Chen S, Jensen GJ. Molecular organization of Gram-negative peptidoglycan. Proc Natl Acad Sci. 2008;105:18953–18957. doi: 10.1073/pnas.0808035105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harwood CR, Cutting SM. Molecular biological methods for Bacillus. Wiley; New York: 1990. [Google Scholar]
- Herbold DR, Glaser L. Interaction of N-acetylmuramic acid L-alanine amidase with cell wall polymers. J Biol Chem. 1975;250:7231–7238. [PubMed] [Google Scholar]
- Holtje JV. Growth of the stress-bearing and shape-maintaining murein sacculus of Escherichia coli. Microbiol Mol Biol Rev. 1998;62:181–203. doi: 10.1128/mmbr.62.1.181-203.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtje JV, Mirelman D, Sharon N, Schwarz U. Novel type of murein transglycosylase in Escherichia coli. J Bacteriol. 1975;124:1067–1076. doi: 10.1128/jb.124.3.1067-1076.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Insall RH, Machesky LM. Actin dynamics at the leading edge: From simple machinery to complex networks. Dev Cell. 2009;17:310–322. doi: 10.1016/j.devcel.2009.08.012. [DOI] [PubMed] [Google Scholar]
- Kuroda A, Rashid MH, Sekiguchi J. Molecular cloning and sequencing of the upstream region of the major Bacillus subtilis autolysin gene: A modifier protein exhibiting sequence homology to the major autolysin and the spoIID product. J Gen Microbiol. 1992;138:1067–1076. doi: 10.1099/00221287-138-6-1067. [DOI] [PubMed] [Google Scholar]
- Lindow JC, Kuwano M, Moriya S, Grossman AD. Subcellular localization of the Bacillus subtilis structural maintenance of chromosomes (SMC) protein. Mol Microbiol. 2002;46:997–1009. doi: 10.1046/j.1365-2958.2002.03235.x. [DOI] [PubMed] [Google Scholar]
- Lopez-Diaz I, Clarke S, Mandelstam J. spoIID operon of Bacillus subtilis: Cloning and sequence. J Gen Microbiol. 1986;132:341–354. doi: 10.1099/00221287-132-2-341. [DOI] [PubMed] [Google Scholar]
- McPherson DC, Popham DL. Peptidoglycan synthesis in the absence of class A penicillin-binding proteins in Bacillus subtilis. J Bacteriol. 2003;185:1423–1431. doi: 10.1128/JB.185.4.1423-1431.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, Garsin DA, Losick R. Self-reinforcing activation of a cell-specific transcription factor by proteolysis of an anti-σ factor in B. subtilis. Mol Cell. 2001;8:873–883. doi: 10.1016/s1097-2765(01)00362-8. [DOI] [PubMed] [Google Scholar]
- Schaeffer P, Millet J, Aubert JP. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci. 1965;54:704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shah IM, Laaberki MH, Popham DL, Dworkin J. A eukaryotic-like Ser/Thr kinase signals bacteria to exit dormancy in response to peptidoglycan fragments. Cell. 2008;135:486–496. doi: 10.1016/j.cell.2008.08.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith K, Bayer ME, Youngman P. Physical and functional characterization of the Bacillus subtilis spoIIM gene. J Bacteriol. 1993;175:3607–3617. doi: 10.1128/jb.175.11.3607-3617.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P, Losick R. Molecular genetics of sporulation in Bacillus subtilis. Annu Rev Genet. 1996;30:297–341. doi: 10.1146/annurev.genet.30.1.297. [DOI] [PubMed] [Google Scholar]
- Uehara T, Park JT. An anhydro-N-acetylmuramyl-L-alanine amidase with broad specificity tethered to the outer membrane of Escherichia coli. J Bacteriol. 2007;189:5634–5641. doi: 10.1128/JB.00446-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uehara T, Dinh T, Bernhardt TG. LytM-domain factors are required for daughter cell separation and rapid ampicillin-induced lysis in Escherichia coli. J Bacteriol. 2009;191:5094–5107. doi: 10.1128/JB.00505-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Straaten KE, Barends TR, Dijkstra BW, Thunnissen AM. Structure of Escherichia coli lytic transglycosylase MltA with bound chitohexaose: Implications for peptidoglycan binding and cleavage. J Biol Chem. 2007;282:21197–21205. doi: 10.1074/jbc.M701818200. [DOI] [PubMed] [Google Scholar]
- Vollmer W, Joris B, Charlier P, Foster S. Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol Rev. 2008;32:259–286. doi: 10.1111/j.1574-6976.2007.00099.x. [DOI] [PubMed] [Google Scholar]
- Youngman PJ, Perkins JB, Losick R. Genetic transposition and insertional mutagenesis in Bacillus subtilis with Streptococcus faecalis transposon Tn917. Proc Natl Acad Sci. 1983;80:2305–2309. doi: 10.1073/pnas.80.8.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou R, Chen S, Recsei P. A dye release assay for determination of lysostaphin activity. Anal Biochem. 1988;171:141–144. doi: 10.1016/0003-2697(88)90134-0. [DOI] [PubMed] [Google Scholar]