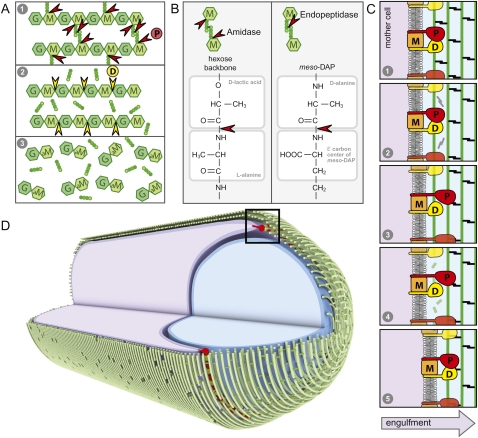
Figure 7.
Coordinated activities in the engulfment complex drive membrane migration around the forespore. (A) Schematic representation of the complementary and sequential activities of IIP and IID. (1) In the diagram, IIP cleaves the stem peptide from the glycan strand and the cross-links between the tetrapeptides. (2) Following peptide cleavage by IIP, IID cleaves the denuded glycan strands into anhydrodisaccharides. (B) Comparison of the amide bonds cleaved by IIP. (C) Schematic diagram of the proposed catalytic cycle of the engulfment complex. (1) IIP (P) and IID (D) in complex with IIM (M) bind the PG. (2) IID stimulates the amidase activity of IIP, resulting in cleavage of the stem peptides and peptide cross-links (black). (3) IIP is released from the PG and rebinds at a nearby site. (4) IID cleaves the denuded glycan strands (green). (5) IID is released from the PG and rebinds adjacent to IIP. The leading edge of the engulfing mother cell membrane (dark purple) moves toward the forespore pole (to the right). (D) Circumferentially distributed engulfment complexes drive membrane movement around the forespore. Shown is a schematic diagram of a sporangium during the morphological processes of engulfment. The membranes of the mother cell (purple) are migrating around the forespore (blue). The glycan strands (green) are shown running perpendicular to the long axis of the cell, as has been proposed for E. coli and C. crescentus (Holtje 1998; Gan et al. 2008). The hoops of glycan strands are stitched together by the cross-linked tetrapeptides. IID (yellow) and IIP (red) are anchored in the leading edge of the mother cell membrane. Processive degradation of the PG drives the mother cell membranes toward the cell pole. For simplicity, we show the engulfment complex acting on the basement layer of the PG meshwork. It is possible that the complex degrades more than one layer, and not necessarily the layer adjacent to the forespore.