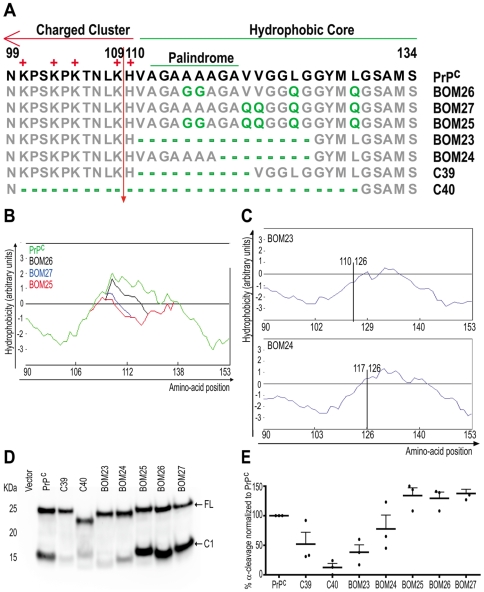
Figure 3. Evaluation of the role of the hydrophobicity in the α-cleavage site region in modulating α-cleavage.
A: Amino acid sequence alignment of the PrPC constructs used. Numbers represent amino-acid residues. The positive residues of the charge cluster, the hydrophobic core, and the palindrome are highlighted. Arrow: α-cleavage site. Grey: non-mutated residues. Green: deletions or mutations into non-charged residues. B: Superimposed hydrophobicity plots of the region 90–153 of PrPC (green) and mutants BOM26 (black), BOM27 (blue), and BOM25 (red). C: Hydrophobicity plot of the region 90–153 of BOM23 in the upper panel and BOM24 in the lower panel. Vertical line represents the site of the deletion spanning the residues 111–125 and 118–125 for BOM23 and BOM24 respectively. Numbers adjacent to the vertical line indicate the amino acid residues flanking the deletion of the mutants. D: Western blot of PNGase treated cell lysates of Hpl cells transfected with the various PrPC deletion constructs used in the current study. Detection was done with POM1. Arrows point to the full length PrP (FL) and C1 fragment. E: Quantification of the percentage of α-cleavage of the various PrPC mutants, based on densitometry of western blots, from which (D) is representative. Quantifications were calculated in the linear range of the densitometric signal of independent experiments. Values refer to the amount of C1 generation comparing to total abundance of PrPC, and are normalized to cleavage of non-mutated PrPC, which was assessed in the same blot. Error bars represent the SEM.