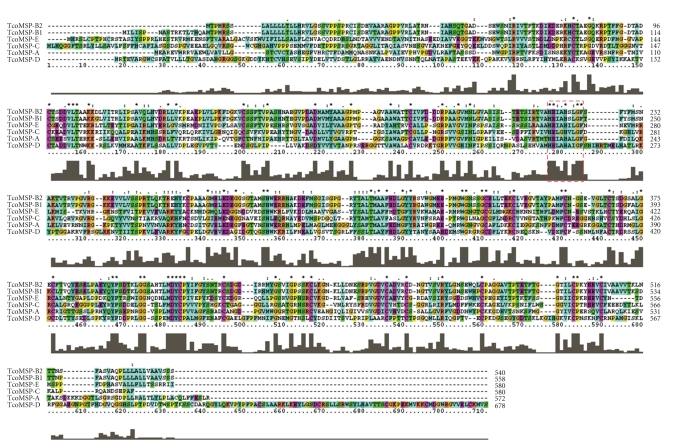
Figure 1.
Multiple sequence alignment of T. congolense MSP homologues. The predicted amino acid sequences for each T. congolense MSP homologue were aligned to each other using the ClustalW alignment tool. Output was directly generated using ClustalX for Windows. The dashed red box indicates a conserved motif shown to be essential for Zn2+ binding in related proteins.