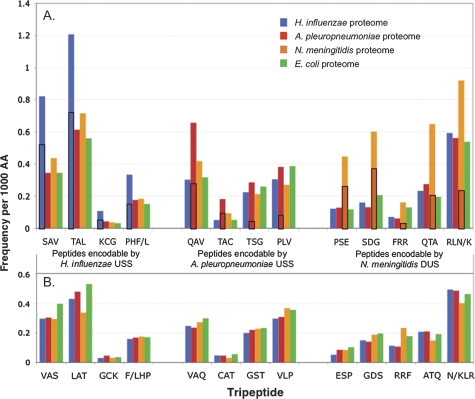
FIG. 1.—
Frequencies of uptake sequence-encodable tripeptides in bacterial proteomes. H. influenzae proteome, blue; A. pleuropneumoniae proteome, red; N. meningitidis proteome, orange; Escherichia coli proteome, green. (A) Tripeptides that can be specified by each of the three uptake sequences: left group by H. influenzae USS; center group by A. pleuropneumoniae USS; and right group by N. meningitidis DUS. The enrichment in the H. influenzae proteome of peptides encodable by the H. influenzae USS was significant (P = 0.010), the enrichment in the A. pleuropneumoniae proteome of peptides encodable by the A. pleuropneumoniae USS was just above the significance threshold (P = 0.058), and the enrichment in the N. meningitidis proteome of peptides encodable by the N. meningitidis DUS was highly significant (P < 0.0001). Boxed areas of bars indicate the fraction of peptides specified by perfect-core uptake sequences. (B) Reversed-sequence tripeptide controls. None of the controls showed significant enrichment.