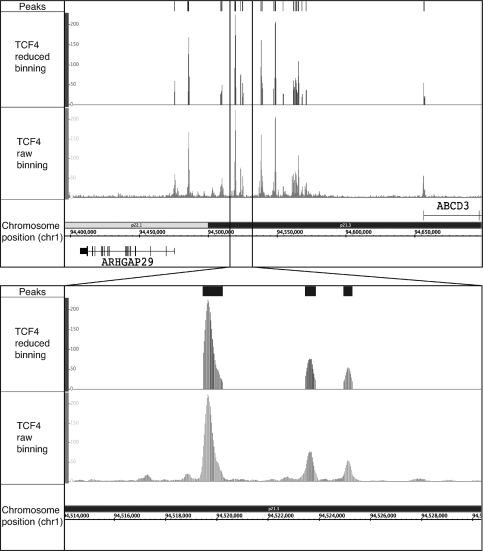
Figure 2.
Visualization of ChIP-seq data using Sole-Search output files. ChIP-seq data for the merged TCF4 replicate dataset were analyzed using Sole-Search (Table 1). Shown for a region of chromosome 1 is: (Peaks) the visualization file (signifpeaks.gff) indicating the called peaks; (TCF4 reduced binning) the visualization file (redbin.sgr) of the tags that correspond only to regions called as peaks, and (TCF4 raw binning) the visualization file (rawbinning.sgr) of all binned and mapped tags. The inset shows the same files, but with an expanded view of a region of chromosome 1. The rawbinning.sgr files are provided for each individual chromosome, due to their large size (e.g. the size of the TCF4 chromosome 1 rawbinning.sgr file is 64.1 MB). However, the redbin.sgr file and the signifpeaks.gff file are much smaller (e.g. the size of the TCF4 redbin.sgr file, which shows TCF4 peaks for all chromosomes, is only 4.5 Mb) and are provided as single files for the entire genome, for ease in comparing different datasets.