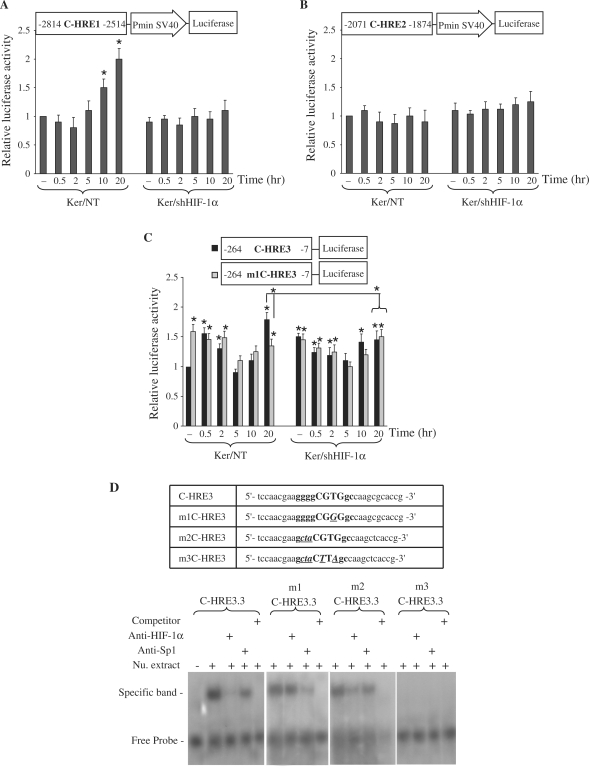
Figure 3.
Functional analysis of putative HREs present in the XPC promoter. Luciferase reporter gene constructs containing C-HRE1 (A), C-HRE2 (B) or C-HRE3 (C) fragments were transfected into non-transduced or shHIF-1α-transduced keratinocytes. Cells were harvested at indicated time points after UVB irradiation. Firefly luciferase activity was normalized to Renilla luciferase activity. The luciferase assays were done in triplicate. For each reporter, the mean ± SD luciferase activity is shown (n = 3) relative to the activity in non-transduced cells prior to irradiation. The schematic representation of the plasmid construct used in each experiment is shown at the top of each diagram. *P < 0.05 for cells at the indicated time point versus NT cells prior to irradiation. (D) EMSA shows that HIF-1α- and Sp1-binding sites on C-HRE3 in XPC promoter are functional. The oligonucleotide sequences used in EMSA are shown in the top panel. C-HRE3 corresponds to the wild-type probe, m1C-HRE3 contains a mutation in the HIF-1α binding site, m2C-HRE3 is a probe mutated in the Sp1-binding site, and m3C-HRE3 contains mutations in both sites. Uppercase letters indicate the core of the HIF-1α binding site, and bold letters indicate the Sp1 binding site that shares a common sequence with C-HRE3.3. Mutated bases are indicated with bold, italics and underlining. Nu. extract, nuclear extract.