Abstract
The daunting task required of the gut-barrier to prevent luminal pathogens and harmful substances from entering into the internal milieu and yet promoting digestion and absorption of nutrients requires an exquisite degree of coordination between the different architectural units of this barrier. The complex integration and execution of these functions are superbly carried out by the intestinal mucosal (IM) surface. Exposed to trillions of luminal microbes, the IM averts threats by signaling to the innate immune system, through pattern recognition receptors (PRR), to respond to the commensal bacteria by developing tolerance (hyporesponsiveness) towards them. This system also acts by protecting against pathogens by elaborating and releasing protective peptides, cytokines, chemokines, and phagocytic cells. The IM is constantly sampling luminal contents and making molecular adjustments at its frontier. This article describes the topography of the IM and the mechanisms of molecular adjustments that protect the internal milieu, and also describes the role of the microbiota in achieving this goal.
1. Introduction
The single-cell epithelial layer of the intestinal mucosa (IM) confronts the largest antigenic microbial challenge of any other mucosal surface in the human body [1]. The presence of large microbial communities in the lumen is a huge problem for the intestinal immune system [2, 3]. In humans, extensive in utero development of T and B cells takes place, and after birth there is a rapid and massive intestinal colonization of antigenic microbes, ultimately establishing stable microbial communities lasting life-long for the individual host [1, 4, 5].
The molecular mechanisms that are involved in shaping and selecting a stable microbiota for different individuals are areas of considerable research interest [6, 7]. There is evidence emerging that the microbiota modulates a series of processes that result in maturation, differentiation, and proliferation of the IM at both cellular and molecular levels [3, 5, 8]. Through a molecular chain of events, the microbiota provides a major drive for the maturation of the innate and adaptive immune systems. It has a profound effect on the intestinal barrier and on distant organs. In this review, we present molecular microbial-mucosal interactions by providing a summary of the different strata of the IM, beginning with the microbiota as the outermost layer, and describe how different layers of the IM form a physical and immune barrier influenced by the microbiota [2, 4, 5, 8]. In this overview, we also describe how microbiota modulates innate and adaptive immunities.
2. Microbial-Mucosal Interactions
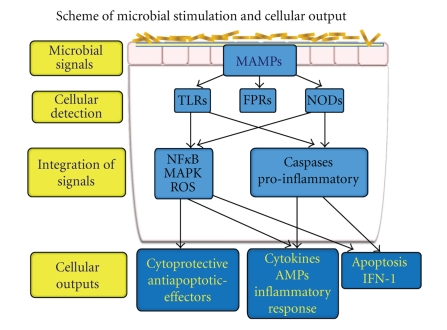
Through a process of “cross talk” with the mucosal immune system, the microbiota negotiates mutual growth, survival, and inflammatory control of the intestinal ecosystem [9, 10]. The IM is equipped with trans-membrane or intra-cytoplasmic receptors, referred to as pattern recognition receptors (PRRs) that are defined by their ability to specifically recognize and bind distinctive microbial macromolecular ligands (Figure 1). These ligands referred to as microbial-associated molecular patterns (MAMPs) include lipopolysaccharide (LPS-a component of gram-negative bacteria), flagellin, peptidoglycans, and formylated peptides [3, 11–14] (Table 1). The transmembrane PRRs include the family of Toll-like receptors (TLRs), that sample the extracellular and endosomal compartments, and the intracellular NOD (Nucleotide-binding oligomerization domain)-like receptors (NLRs) that confront and protect the cytoplasmic compartment [11–14]. This microbial-mucosal intersignaling cultivates immune tolerance (hyporesponsiveness) resulting in the development of a stable core microbiota. Establishing a core microbiota of diverse and native commensal species is critical and advantageous to the host as it provides competition to the pathogenic microbes [9, 15]. This process prevents pathogens from forming a niche for their persistence and proliferation [16]. In response to viral components, signaling by TLRs results in expression of Interferon 1 (IFN1). For fungal components, C-type lectin receptors serve as PRRs [13]. On the surface of neutrophils, trans-membrane receptors belonging to the family of formylated peptide receptors (FPRs) are present as high affinity PRRs for MAMPs [17]. Upon exposure to MAMPs, signals from FPRs activate neutrophil transduction pathways to generate reduced NADPH (nicotinamide adenine dinucleotide phosphate) oxide (NOX)-dependent reactive oxygen species and promote phagocytic motility [17]. Recently, several neutrophil FPRs have been characterized in intestinal epithelial cells (IECs), suggesting that these epithelial receptors may mediate microbial monitoring in the gut in a manner analogous to their traditional functions in phagocytes [18]. Disruption in the ligand process of PRRs with MAMPs has been linked with inflammatory intestinal diseases [3]. One such example is Crohn's disease, in which mutant forms of NLR NOD2 have been identified [3, 19, 20].
Figure 1.
Commensal microbiota, or pathogens, at the mucosal surface create signals, called microbial-associated molecular patterns (MAMPs), to stimulate pattern recognition receptors (PRRs) including toll-like receptors (TLRs), formylated peptide receptors (FPRs), or nucleotide-binding oligomerization domain-like receptors (NODs). Subsequent signaling consists of an intricate and inter-relational pathway, which determines the signaling output based on the initial perception of the triggering organism. Output can be a protective response to commensal microbiota, an inflammatory response to pathogenic organisms, or triggers for apoptosis.
Table 1.
Pattern recognition receptors (PRRs), microbial-associated molecular patterns (MAMPs), and expression patterns (see, [3, 12, 14]).
| PRRs | Location | MAMPs | MAMPs |
|---|---|---|---|
| Non Viral Ligands | Viral Ligands | ||
| TLRs | |||
| TLR1 | Cell membrane | Lipopeptides | |
| TLR2 | Cell membrane | Di-/triacyl lipopeptide | Virion (HSV) |
| Peptidoglycan | HA (Measles) | ||
| Lipoteichoic acid (LTA) | |||
| Zymosan | |||
| Porins | B and H proteins (HCMV) | ||
| Lipoarabinomannan | |||
| Phospholipomannan | ENv (MMTV) | ||
| Glucuronoxylomannan | |||
| GPI-linked proteins | Core, NS3 (HCV) | ||
| Virion | |||
| TLR3 | Endosome membrane | dsRNA | RNA viruses |
| LR4 | Cell membrane | Lipopolysaccharide (LPS) | ENv protein (MMTV) |
| Mannan | |||
| Glucuronoxylomannan | F protein (RSV) | ||
| HSP | |||
| Fibrinogen | |||
| TLR5 | Cell membrane | Flagellin | |
| TLR6 | Cell membrane | Lipoprotein, LTA, others | |
| TLR7 | Endosome membrane | Synthetic ssRNA (e.g., imiquimod, resiquimod (R848) | Influenza A, VSV |
| TLR8 | Endosome membrane | Synthetic ssRNA (e.g.,imiquimod, resiquimod (R848) | HIV |
| TLR9 | Endosome membrane | CpG DNA | Unmethylated DNA (HSV 1,2) |
| Hemozoin | MCMV | ||
| Adenovirus | |||
| Baculovirus (wild type) | |||
| TLR10 | Cell membrane | Unknown | |
| TLR11 | Cell membrane | Profilin | |
| NLRs | |||
| Nod1 | Cytoplasmic | Gram-negative peptidoglycan | |
| Nod2 | Cytoplasmic | Gram-negative and positive peptidoglycan | |
| IPAF | Cytoplasmic | Flagellin | Viral RNA |
| NALP3 | Cytoplasmic | RNA | |
| FPR | Cell membrane | Formylated peptides, ? | |
| FPRL 1-2 | Cell membrane | Formylated peptides, ? | |
| RIG-1 helicase | Cytoplasmic | Synthetic dsRNA | HCV, |
| 5′-triphosphate | Japanese encephalitis | ||
| dsRNA | RSV, Influenza A, EBV | ||
| C-type lectins | Cell membranes | Fungal carbohydrates | |
Abbreviations: dsRNA: double-stranded RNA; EBV: Epstein-Barr virus; ECMV: encephalomyocarditis virus; GPI: glycosylphosphatidylinositol; HA: hemagglutinin; HCV: hepatitis C virus; HCMV: human cytomegalovirus; HIV: human immunodeficiency virus; HSP: heat-shock protein; HSV: herpes simplex virus; LPS: lipopolysaccharide; MCMV: murine cytomegalovirus; MMTV: mouse mammary tumor virus; RSV: respiratory syncytial virus; ssRNA: single-stranded RNA; VSV: vesicular stomatitis virus.
2.1. Regulatory Pathways
In the IM, activation of PRRs initiates regulatory pathways such as nuclear factor κB (NFκB)/Rel pathways, mitogen-activated protein kinase (MAPK), and caspase-dependent signaling cascades [14, 19–21] (Figure 1). The transcriptional response of the IM to microorganisms can be different. Many PRR ligands are expressed by commensal bacteria yet the IM does not activate an inflammatory response to these bacteria. Conversely, some commensal bacteria exert protective effects by attenuating proinflammatory responses induced by pathogenic bacteria. Inflammatory or apoptotic responses to pathogenic bacteria, or stress signals, are controlled by NFκB and caspase-dependent signaling [3, 21–23]. Most commensal organisms limit the signaling of NFκB by inhibiting epithelial proteosome function, inhibiting degradation of the IκB (the counter regulatory factor to NFκB), or by exporting the NFκB subunit p65 out of the nucleus through a peroxisome proliferator-activated receptor (PPAR)γ-dependent pathway. Induction of transforming growth factor-β (TGF-β) and MMP3K pathways has also been implicated in anti-inflammatory and antiapoptotic effects mediated by commensals [3, 24–28]. The TLRs can also initiate proapoptotic signaling. The bacterial lipoprotein-TLR2 interactions activate the caspase-8 pathway of apoptosis via the myeloid differentiation primary-response gene 88 (Myd88) and by subsequent recruitment of fast activated death domain (FADD) pathways. Recent findings utilizing purified flagellin and flagellate/aflagellate bacteria in both in vitro and in vivo systems have shown that important roles are played by flagellin in the modulation of apoptotic responses [21]. In response to cellular (IEC) attack by flagellate pathogens, signal transduction through TLR5 initiates proinflammatory transcriptional responses. Macrophages utilize intracellular IL-1β-converting enzyme protease-activator factor (IPAF)/neuronal apoptosis inhibitory protein (NAIP) 5 to detect intracytoplasmic flagellin and respond with IL-1 release and/or apoptosis. Thus, this interaction between TLR5 and flagellin initiates pro-inflammatory and pro-apoptotic pathways. If pro-inflammatory signaling is unimpeded, transcriptional activation of a battery of anti-apoptotic/cytoprotective genes arrests the apoptotic pathways and allows inflammation without cell death [21]. Analogous to these events, it has been reported that during infection with wild type Salmonella, flagellin-induced pro-inflammatory signaling reduces cellular pro-apoptotic responses to bacterial effectors, whereas the absence of this potent pro-inflammatory determinant in mutants allows unhindered apoptosis. In this case, activation of pro-inflammatory signals protects apoptosis. Thus, the complex process of inflammation is intertwined with the process of apoptosis [21].
When activated, the interactive mucosal-microbial information system relays posttranslational events to the transducer PRR which in turn transmits the message, through transcriptional or post-transcriptional processes, to the effector cell [1, 5, 22–25].
3. Barrier Defense and Innate Immune System
3.1. Microbiota
By adulthood, a vast consortium of microbiota are supported in the intestinal lumen, but with current microbiologic methods, less than 30% of those microbes are culturable [15, 29]. Despite our lack of understanding of the true diversity of luminal microbes, new molecular approaches are allowing census-based culture-independent inventories of the entire microbiota and defining the microbial taxonomy utilizing genomic technology [11, 30]. The most common approach to generate DNA sequence data is to amplify the genes encoding RNA in small ribosomal subunits (16S rRNA) using primers targeting generally conserved regions of the gene. Rapid and cost-effective methods that provide a “fingerprint” of the microbial diversity in the individual components of the microbota are then characterized by cloning and DNA sequencing [15, 30–32].
Animal experiments and comparisons between conventionally raised mice (or rats) with germ-free counterparts have revealed that a series of anatomic, biochemical, and physiologic functions are performed by the microbiota that are crucial for development, maintaining the integrity of the barrier function, and for the repair of the IM [5]. The fact that so many morphological intestinal tissue defects appear in germ-free animals indicates that the development of the IM is inherently connected to microbial luminal colonization. For example, in contrast with conventionally raised mice, the villus capillaries in germ-free mice develop poorly during weaning and remain poorly developed until adulthood, indicating a microbial contribution to angiogenesis of the villus-core. This was further confirmed when it was shown that bacterial colonization of germ-free mice resulted in rapid and dramatic reinduction of angiogenesis [5, 33]. Electrophysiologic studies indicate that microbial colonization improves intestinal motility and modulation of enzyme activity [34]. Several important effects of the microbiota on the development of the immune system have been ascertained by selectively colonizing germ-free animals and then evaluating immune responses that have not been influenced by any other microbial molecules [11, 28, 35]. Germ-free animals show extensive defects in the development of gut-associated lymphoid tissue (GALT) and antibody production, fewer and less cellular lymphoid follicles (Peyer's patches), a thinner and less cellular lamina propria, and fewer plasma cells in germinal centers of the mesenteric lymph nodes compared with animals housed under conventional conditions [28, 36, 37]. These findings indicate that the development of the ultrastructure of the IM is dependent on luminal bacteria. The microbiota also influences the morphology of the IEC. Germ-free mice have been shown to develop an altered morphology of their microvilli and a reduced rate of turnover of their IECs (intestinal restitution) compared to conventionally raised animals [38]. Furthermore, the mircobiota has been shown to direct the glycosylation of surface proteins of the IEC [39].
The microbiota also contributes to the development of intra-epithelial lymphocytes (IEL) as evident by the fact that numbers of αβ T-cell receptor (TCR)-bearing IELs (αβIELs) are reduced in germ-free mice compared to conventionally raised mice and that γδ TCR-bearing IELs (γδIELs) isolated from conventionally raised mice are cytolytic but not those isolated from germ-free mice [5, 40–45]. These findings indicate that the microbiota influences maturation and execution of several immune functions that benefit the host IM. In return, after birth, changes occur in the host IM that promote colonization of commensal organisms. Conventionally raised mice begin life by expressing intestinal epithelial glycans that predominantly have sialic acid as their terminal moiety. During weaning there is a shift towards terminal fucose that does not occur in germ-free mice [40]. Fucosylation of glycans facilitates colonization by commensal species, such as B. thetaiotaomicron, that use terminal fucose as an energy source [46]. Thus, the host's IM encourages colonization of commensal organisms by shifting its energy source in their favor so that the commensals can gain control over the pathogenic species in the competitive intestinal ecosystem. Once established, the microbiota then shape their niche in a way that promotes morphological modulation of the host's IM in favor of that host [5, 8]. Another example is that, compared to conventionally raised mice, germ-free mice never achieve high activity of Angiogenin-4 (Ang4), a Paneth cell protein with potent bactericidal activity that plays important role in epithelial cell defense during the postweaning-period in mice, indicating that induction of Ang4 in Paneth cells of the IM is regulated by the microbiota [43].
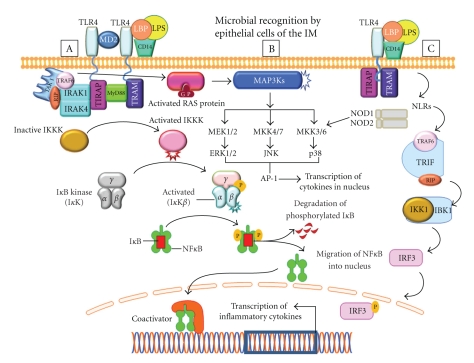
The microbiota regulates the intestinal innate immune system by modulating expression of TLRs and NOD/CARD (caspase recruitment domain) mediated activation of immunosensory cells through MAMPs [27]. Decreased cytoprotective factors and decreased enterocyte proliferation have been observed in TLR-defective mice [47, 48]. Individual members of the microbiota can dampen TLR-mediated inflammatory signals and exert protective effects by attenuating pro-inflammatory responses [27, 49] (Figure 1). Several studies have shown how commensals, and those bacteria with probiotic function, can suppress inflammatory signals [50, 51]. One of the components of regulation of inflammatory signals is by activating IκB, the inhibitory component of NFκB activation. Manipulation of the NFκB pathways has revealed both anti-inflammatory and pro-inflammatory roles for this transcriptional control pathway, suggesting temporal patterns in which TLR and NFκB pathways are activated in response to distinct microbial signals. Mice with IEC-specific knockout genes encoding for Iκκγ or Iκκβ, the 2 components of the inhibitory κB kinase (Iκκ) complex that activates NFκB, were found to be susceptible to chemically induced colitis or to spontaneous development of intestinal inflammation, respectively [50–53]. Upon binding to their respective MAMPs, the TLRs act together with the Myd88 adapter protein to induce intracellular signaling events that converge upon the NFκB and MAPK pathways to regulate the expression of genes that are involved in commensal-induced fortification of the IM and pathogen-induced inflammation (Figures 1 and 2). TLR ligation triggers recruitment of Myd88 to the receptor complex through TIR-TIR domain interactions. The death domain of Myd88 recruits a death-domain containing protein known as IL-1-R associated protein kinase (IRAK). Activation of IRAK leads to activation of NFκB, p38 MAPK, and other regulators of gene expression. These events and expression of pro-inflammatory genes form the basis of innate immune response. Studies demonstrating loss of Myd88 function in dendritic cells established that TLR/Myd88-mediated MAMP recognition activates dendritic cells to produce pro-inflammatory cytokines and promotes T-helper responses [41, 49] (Figure 2).
Figure 2.
A schematic illustration of the recognition of microbial associated molecular patterns (MAMPs), such as LPS (lipopolysaccharide), by pattern recognition receptors (PRRs) on epithelial cells and activation of three (A, B, and C) pathways that induce production of pro-inflammatory cytokines or chemokines. A. Expressed on the cell membrane of most intestinal epithelial cells (IECs), toll-like receptors (TLRs) are triggered by LPS. Four toll-like interleukin receptor (TIR) adaptors become involved in propagating TLR signaling, including Myd88 (myeloid differentiation primary-response gene 88), TIRAP (toll-interleukin-1 receptor domain containing adapter protein), TRAM (translocating chain-associated membrane protein), and the TRAF (TNF receptor associated factor) protein family. This interaction induces phosphorylation and activation of TAK1, leading to activation of IKKs. Inactive IKK sequesters NFκB in the cytoplasm and leads to degradation of IκB, with subsequent release of NFκB that induces transcription of pro-inflammatory cytokines and chemokines. B. In various cells, Myd88-dependent signaling is associated with activations of TAK1, also known as MAP kinases. Extracellular signal-regulated kinase (MEK) is an intermediary of the MAPK pathway. Activated MEK subsequently phosphorylates ERK which translocates to the nucleus where it activates multiple transcription factors. Cytoplasmic nucleotide-binding oligomerization domain-like (NOD) receptors also recognize MAMP. The ligand NOD1 recognizes a peptidoglycan, a constituent only of gram-negative bacteria. NOD2 recognizes constituents of both gram-positive and gram-negative bacteria. NODs transduce signals in the pathway of NFκB and MAP kinases.
In another study, IEC-specific deletion of ReIA/P65, which encodes the primary subunit of the NFκB transcription factor, increased epithelial cell proliferation and caused apoptosis and increased susceptibility to colitis. These findings indicate that NFκB activation through TLRs on the IM promoted antinflammatory responses to microbial signals and promote innate immunity [10–14, 16, 20, 24, 27].
Uncontrolled signaling through TLRs can potentially cause excessive inflammation. Intestinal epithelial cells constitutively, or inducibly, express high levels of the TLR inhibitor Toll-interacting protein (TOLLIP) [11, 21, 26]. Expression of TOLLIP has been shown to correlate with the in vivo luminal bacterial load and is highest in healthy colonic mucosa [26]. There are other regulators, including single immunoglobulin IL-1R-related protein (SIGIRR), IRAK-M, A20, PPARγ, and NOD2, which exert inhibitory influence over inflammation. Evidence suggests that similar to TOLLIP, NOD2 might suppress the inflammatory cascade and mutations of NOD2 that are associated with Crohn's disease. These inhibitory molecules are important in maintaining microbial homeostasis [26, 28, 35, 40].
The microbiota also functions as a regulator of nutrient metabolism, allowing the host to digest many nutrients that would otherwise be inaccessible to the IM [3, 25–27, 36]. The complex carbohydrates that would otherwise be indigestible are degraded into short-chain fatty acids (SCFA) and monosaccharides by the mircobiota. These SCFA serve as a source of energy and regulate growth and differentiation of IECs, especially in the colon. The microbiota also degrades mucus glycoproteins and maintains the specific landscape of the IM. Germ-free animals exhibit a dramatic enlargement of the cecum, largely due to accumulation of undegraded mucus. In addition, the microbiota modifies the differentiation programs of intestinal epithelial lineages at key points during their morphogenesis [5].
3.2. Mucin
A coating of mucus on the IM forms the front line of defense segregating virulent organisms and protecting against acidic gastric and duodenal secretions [54, 55] (Figure 3). This mucus gel consists predominantly of high molecular weight glycoproteins (mucins) imparting characteristic polymeric, viscoelastic, and protective properties. Mucins have a high negative surface charge and a large hydration capacity. This mesh-like mucin gel impedes diffusion of offending macromolecules and yet is able to perform functions such as lubrication for the passage of particulate nutrients, maintenance of epithelial hydration, and facilitation of the exchange of gases and nutrients across the IM. So far 21 different mucin genes have been identified. In the intestine, MUC2 is the major mucin produced by goblet cells of the IM. Recent investigations in mice colon have demonstrated that there are two mucus layers of MUC2, an inner mucus layer that is adherent to epithelial layer and an overlying loose mucus layer. The inner layer is densely packed. It is devoid of bacteria suggesting small pore size that physically prevents bacterial penetration [53]. Conversely, the outer loose layer contains high number of bacteria. Because the mucin and protein composition of both the firm and loose layers are identical, it appears that firm layer separates to form loose layers. In small intestine, the same MUC2 is not adherent to epithelial layer but forms the mucus gel that is synthesized in goblet cells. Following synthesis in goblet cells, mucins are packaged in granules, transported to the cell surface, and secreted into the intestinal lumen. Mucins can be secretory or membrane-bound [53–55].
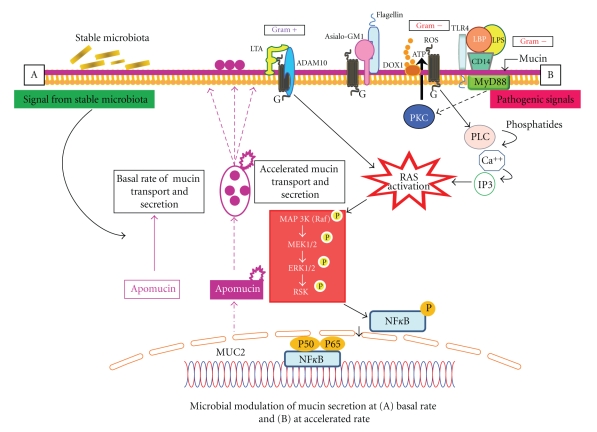
Figure 3.
Under normal physiological conditions, mucin secretion occurring at a stable rate (A) with signals from stable (commensal) microbiota. Mucin secretion is accelerated (B) upon activation by pathogenic organisms. In gram negative bacteria (e.g., Pseudomonas aeruginosa), both LPS (lipopolyscaccharide) and flagellin signal activation of RAS-MEK1/2-ERK1/2 (reticulo-activating system-pathway—meiosis-specific serine/threonine protein kinase-extracellular signal-regulated kinase) pathways. Bound to LPS binding protein (LBP), LPS uses a toll-like receptor (TLR). Flagellin binds to a glycolipid receptor, Asialo-GM1, which is calcium dependent. Lipoteichoic acid (LTA), a component of gram positive bacteria, binds to platelet activating factor (PAF) and activates ADAM10 (adhesion and protease domain molecule) which engages RAS/RAF/MEK/ERK/pp90RSK/NFκB and MUC2 (intestinal mucin) production.
3.2.1. Mucin and Innate Host Defense
Under normal conditions, microbes localize in the mucus, sharing either “self” signature molecules or pathogen-associated molecular patterns. These patterns are in turn recognized by TLRs or other PRRs. Recognition of LPS is achieved through the combined action of membrane-bound or soluble mucin, LPS-binding protein (LBP), CD14, and TLR4 [55] (Figure 3).
In gram-negative bacteria (e.g., Pseudomonas aeruginosa), both LPS and flagellin play a role in altering mucin production (Figure 3). The Binding of LPS to LBP and then to CD14 leads to activation of Ras-MEK1/2-Erk1/2 (reticulo-activating system-pathway—Meiosis-specific serine/threonine protein kinase-extra-cellular signal-regulated kinase) using TLR-4 as a coreceptor. Flagellin, on the other hand, binds to the surface glycolipid receptor Asialo-GM1. The activation of mucin transcription through Asialo-GM1 is calcium dependent as seen by an increase in calcium levels following administration of flagellin. Binding to Asialo-GM1 leads to ATP release and its subsequent binding to the cell surface G protein-coupled receptor (GPCR) [55]. This activates phospholipase C and creates a subsequent increase in intracellular calcium levels. These events finally lead to downstream activation of the Src (rous sarcoma virus-cytoplasmic protein)-dependent Ras pathway, leading to the activation of NFκB and mucin transcription (Figure 3).
Through a different pathway, the gram-positive bacterial product lipoteichoic acid (LTA) binds and activates the platelet-activating factor (PAF) receptor, which is a cell surface G-protein coupled receptor (Figure 3). This leads to activation of ADAM10, which then cleaves the trans-membrane heparin-binding EGF (epidermal growth factor), which in turn activates the EGF receptor. This leads to the engagement of the Ras/Raf/MEK/ERK/pp90rsk/NF-κB pathway and MUC2 (mucin) transcription [54–56].
The defensive ability of mucin lies in its capacity to entrap microbes. Adhesion to specific mucin epitopes is thought to facilitate mucus colonization by commensal bacteria, thereby providing a number of growth advantages. Accordingly, intestinal mucin is thought to dictate the composition of the bacterial community [55] (Figure 3).
3.2.2. Other Goblet Cell Secretions in Innate Host Defense
In addition to mucin production, goblet cells also produce two other innate defense molecules: the intestinal trefoil factor and the resistin-like molecule-β (RELM-β) proteins [55, 56]. There is evidence that these two innate defense molecules may stabilize the mucin polymer and/or regulate mucin secretion [55]. Importantly, these molecules may need a mucin medium to exert their biological functions.
3.2.3. Intestinal Trefoil Factor
This is a small cystein-rich peptide belonging to the family of trefoil factors (TFF) [57]. In humans 3 TFFs have been identified; TFF1, TFF2, and TFF3. Secreted TFFs act on the IM either extracellularly to augment barrier function or intracellularly in transcriptional and signaling events. Trefoils seem to protect the epithelium and heal injured mucosa. With mucosal injury, TFFs are up-regulated and stimulate epithelial restitution [57]. They may also play a role in mucus stabilization by interacting or cross- linking with mucin to aid in the formation of gel. Since TFFs may be coexpressed with secreted mucins, there is possibly a synergistic action in mucosal protection and repair between the two since they are coexpressed in both normal and injured mucosae [55, 57].
3.2.4. Resistin-Like Molecule-β (RELM-β)
This is a member of the resistin-like molecule (RELM) family. RELM- β is expressed in both the small and large intestine within IECs and particularly in goblet cells. RELM-β regulates barrier integrity and susceptibility to inflammation. It appears that although mucin and RELM-β are secreted by goblet cells, RELM-β can also act as an effective luminal secretagogue [55, 58–60].
3.3. Tight Junction (TJ) Assembly
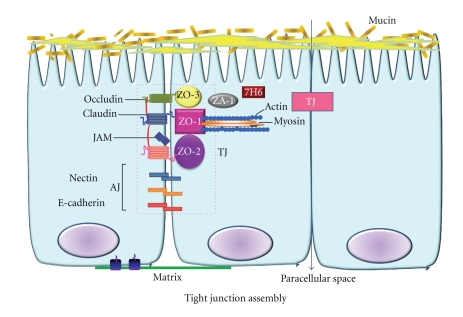
The TJ assembly is a cluster of proteins between intestinal epithelial cells forming an effective barrier between the lumen and lamina propria [61]. In general, there are three types of junctional complexes, the tight junctions (TJs), adherens junctions (AJs) and desmosomes, and gap junctions [61–64]. Of these three, TJs represent the major barrier within paracellular pathways between intestinal epithelial cells [61, 64] (Figure 4). The intestinal TJ-complex-associated proteins consist of intracellular and surface membrane proteins. The intracellular proteins are: zona occludens (ZO)-1, ZO-2, and ZO-3, cingulin, 7H6, symplekin, and ZA-1 [61, 64, 65]. The membrane proteins localized to the TJ are the following: occludin, claudin, junctional adhesion molecules (JAMs), and the coxsackie virus and adenovirus receptor (CAR) proteins [61, 62, 64–68]. Studies suggest that claudin-1 may directly associate with occludin laterally in the membrane within the same cell and the combination of these two proteins functioning together performs the major “gatekeeper” or barrier function of the tight junction [61, 64–69].
Figure 4.
A model of the protein components of tight (TJ) and adherens junctions (AJ) in a highly polarized epithelial cell. The TJ is composed of transmembrane proteins (occludins, claudins, and JAMs) linked to an actin cytoskeleton via cytoplasmic ZO (zonula occludens) proteins. ZO-1 binds to actin. The AJ is composed of the nectin-afadin system and the E-cadherin-catenin system. Nectin's direct partner, afadin, binds to several proteins, including ZO proteins. Thus TJs and AJs may be closely associated, forming the apical junctional complex, which is linked to the actin cytoskeleton network.
Several signaling pathways of the TJ assembly are being investigated. Studies indicate that regulation of TJ assembly occurs through phosphatidylinositol 3-kinase (PI3K) [63]. Prostaglandins have been found to stimulate the recovery of paracellular resistance via a mechanism involving transepithelial osmotic gradients and PI3K-dependent restoration of TJA. Other pathways implicate two Rho family GTPases, Rho and Rac, and light chain-associated myosin protein kinase 20 for regulation of tight junction proteins. The latter has been implicated as a mechanism for stress and cytokine-induced increases in TJ permeability [63].
A number of microorganisms have been shown to attack the intercellular TJ proteins [61]. Enteric pathogens can disrupt the TJ of IECs through a number of different virulence factors. Disruption of the TJ by enteropathogenic Escherichia coli (EPEC) has been attributed, in part, to altering occludin distribution from the TJ into the cytosol. C. difficile toxins A and B have been shown to disorganize apical and basal F-actin and cause dissociation of occludin, ZO-1, and ZO-2 from the lateral TJ membrane [65–68]. Rotavirus infection of IECs has been shown to increase paracellular permeability and cause alteration of F-actin. Furthermore, it has been determined that the NSP4 (nonstructural protein) of rotavirus reduces transepithelial electrical resistance, redistributes filamentous actin, and prevents lateral targeting of the TJ-associated ZO-1 protein [65–68].
An endogenous protein called “zonulin,” which is functionally and immunologically related to zonula-occludin toxin from Vibrio cholera, has been found to disassemble intercellular TJs via interaction with cell membrane receptors [65–68]. It is speculated that dysregulation of zonulin in many diseases may involve loss of cell junction integrity. Serum zonulin is not only up-regulated in celiac disease, but also in type 1 diabetes and multiple sclerosis, suggesting the role for a “leaky gut” in the development of autoimmunity [61].
Despite their complex organization, neither AJs nor TJs are static structures and they can be rapidly disassembled and reorganized in response to various extracellular stimuli. Internalization of AJs and TJs appears to be a common mechanism that rapidly down regulates cell to cell adhesion and allows remodeling of intercellular junctions. Internalization is also induced by various pathophysiologic stimuli including microbial virulence factors, pro-inflammatory cytokines, and oxidative stress [61, 62, 64, 66–69]. Breakdown in the TJA and in the interepithelial barrier associated with intestinal bacteria has been implicated in several diseases including inflammatory bowel disease (IBD), type I diabetes, and obesity [61, 66]. Recent studies indicate that the modulation of microbiota by selectively increasing Bifidobacterium-spp improves barrier function and function of TJA-proteins [66]. Endotoxemia and low-grade inflammation seen in patients with obesity and type I diabetes is believed to be associated with dysbiosis, the presence of a pathogenic microbiota [61, 66, 69]. Developing specific therapeutic strategies to stabilize the microbiota, such as the use of probiotics and prebiotics as an adjunct to management of some of these diseases holds some promise [70].
3.4. Intestinal Epithelial Layer
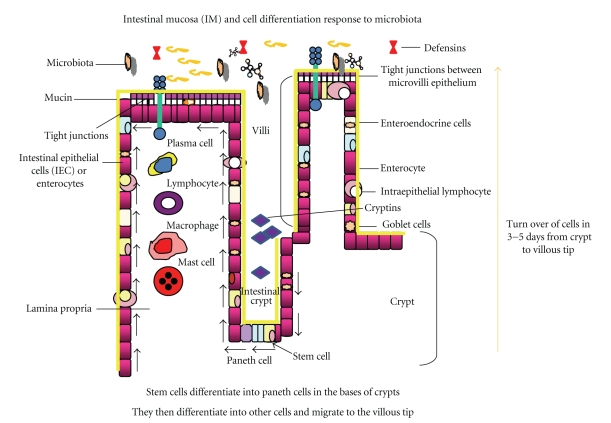
Composed of a single layer of cells that is only ~20 μm thick, the intestinal epithelial layer has evolved strategies to derive maximum nutritional benefit from luminal contents and maintain a beneficial host-microbial relationship while keeping the pro-inflammatory potential of the microbiota under control [36]. As an interface between the host and the environment, the mucosal epithelium consists of well organized crypts and villi supported by microvasculature, as well as lymphatic and connective tissues of the lamina propria and submucosa [1, 25, 27] (Figure 5). Other cellular components include lymphocytes, macrophages, and fibroblasts that perform special functions to maintain the integrity of the IM [1–3, 11, 40, 50, 51, 71].
Figure 5.
A depiction of the intestinal epithelial barrier. The intestinal epithelial barrier consists of a rapidly renewing epithelial cell population, derived from stem cells in the bases of intestinal crypts, that differentiates into other epithelial cell types (Paneth cells, goblet cells, enteroendocrine cells, enterocytes, intra-epithelial lymphocytes, and M cells). These cells, as they are produced in the crypts, migrate to the tips of the villi over approximately 3 to 5 days. This process of repair and restitution can be influenced by cellular signaling pathways initiated by the intestinal microbiota. (Modified from Sharma R, Young C, Mshvildadze M, Neu J. Intestinal Microbiota Does It Play a Role in Diseases of the Neonate? NeoReviews Vol.10 No.4 2009 e166, with permission).
Studies have shown that several immune functions of IECs are influenced by the microbiota, including secretions of cytokines and chemokines and expression of major histocompatibility complex (MHC) molecules that directly interact with lymphocytes [1, 2]. The expression and localization of PRRs (such as TLRs) is also influenced by the microbiota [3, 24, 26, 28, 41, 47, 48]. Expression of several peptides, such as defensins, is defective in germ-free animals [28]. Microbial colonization of germ-free mice induces the production of regenerating islet-derived 3γ (RegIIIγ), a secreted C-type lectin. RegIIIγ is shown to have antimicrobial activity by binding to peptidoglycans, suggesting that microbial species actively shape the intestinal ecosystem [70–72]. In another study, using cocolonization of germ-free mice with B. thetaiotaomicron (a symbiont) and B. longum (a probiotic), Sonnenburg et al. showed that B. longum can increase the diversity of polysaccharides that can be degraded by B. thetaiotaomicron, thus, demonstrating that distinct intestinal bacterial species can affect each other's function [73].
3.4.1. Enteroendocrine Cells (EEC)
More than ten distinct types of EEC have been identified in the adult human [1]. Entero-endocrine cells secrete serotonin, somatostatin, motilin, cholecystokinin (CCK), gastric inhibitory peptide (GIP), neurotensin, vasoactive intestinal peptide (VIP), and enteroglucagon. As EECs migrate to the villus tip the number of cytoplasmic granules increases but the ability to divide decreases. Attempts to map the distribution of EECs using immune-cytochemical techniques in transgenic adult mice have been made [74]. The microbiota has been shown to regulate host energy balance and have an impact on obesity. This effect is mediated through Gpr41, a G protein-coupled receptor expressed by a subset of EECs in the IM [74].
3.4.2. Paneth Cells
These cells are the only cell lineage derived from the crypt stem cells that migrate downward towards the crypt base. Paneth cells produce lysozyme, phospholipase A2, TNF-α, cryptdins, and guanylin [75]. The guanylin family of peptides consists of three endogenous peptides, one of which has a similar primary structure and biological activity as that of Escherichia coli heat-stable enterotoxin (STa). The guanylin and STa both activate intestinal guanylate cyclase-C (GC-C) and elicit 3′–5′ cyclic guanosine monophosphate (cGMP) accumulation in the intestinal mucosa. The activation of cGMP drives secretion of fluids into the intestinal lumen [40, 42, 43, 76].
The cryptdins are defensin-like peptides in intestinal crypts (Figure 5). Cryptdins kill microbes by forming pores in their limiting membranes [40, 75]. At least 17 isoforms have been characterized from a cDNA library. Cryptdin 3 (Cr3) has been shown to induce IL-8 secretion in a dose-dependant manner when applied to a human intestinal cell line (T84) by activating NF-κB and p38 MAPK in a calcium-dependent manner, without influx of extracellular calcium (Ca++). Unlike other known inflammatory agonists, signal transduction by Cr3 occurs slowly, suggesting that selective cryptdins may amplify their roles in innate immunity by acting as paracrine agonists to coordinate an inflammatory response with the antimicrobial secretions of Paneth cells [40, 75]. Because of their ability to kill gram-positive and gram-negative bacteria, fungi, spirochetes, and some enveloped viruses, cryptdins are classified as broad-spectrum antimicrobial peptides [40, 75, 77–79]. Although it is the least expressed of the six isoforms, cryptdin-4 is the most bactericidal. Procryptdins, however, are not bactericidal and thus require degradation of the pro-region by matrix metalloproteinase-7 (MMP-7) for activation. In response to bacterial antigens, Paneth cells release their secretory granules into the lumen of intestinal crypts [40, 79–81].
Defensins are small cationic polypeptides and are the predominant antimicrobial proteins present in a large number of expressed genes [1, 80, 81]. Mammalian defensins are divided into two main structural groups, α and β. α-defensins are particularly plentiful in neutrophils and intestinal paneth cells. The antibacterial activity of defensins is generally ascribed to their ability to disrupt membrane integrity and function, which ultimately leads to the lysis of the microorganism [40]. In addition to exerting direct antimicrobial effects, defensins facilitate and amplify innate and adaptive immune responses, such as activation and de-granulation of mast cells, cytokine production and secretion, maturation of dendritic cells, and chemotaxis of immune cells [81–83].
Integrins are receptors that mediate attachment between IECs and the tissues surrounding them, which may be other cells or the extracellular matrix [40, 82, 84, 85]. Integrins also play a role in cell signaling and thereby define cellular shape and mobility, and regulate the cell cycle. Integrins regulate the assembly of adhesive junctions as well as the activation of various signaling pathways involved with complex organization of the epithelial-cell matrix in maintaining the crypt-villus axis [1, 82–84].
3.4.3. Microfold (M) Cells
These cells are restricted to the dome epithelium overlying the lymphoid follicles of Peyer's Patches in the lamina propria and other lymphoid follicles in the gut [1, 11, 83]. The surface of M cells is characterized by microfolds and the numerous vesicles contained within. These cells perform a significant role in sampling the luminal milieu. They also appear to be the avenue of infection for viral agents (e.g., rotaviruses of the family, reoviridae) [82, 83]. The numerous vesicles appear to facilitate transport of luminal antigens, macromolecules, and microorganisms to the underlying lymphoid tissue [83].
4. Barrier Defense and Adaptive Immune System
Exposed to a vast number of antigens, the IM is protected by a large immune system [1, 2]. Both lymphoid cells (T and B cells) and myeloid cells (macrophages, neutrophils, eosinophils, and mast cells) have a copious presence in the gut [14, 16]. The GALT has evolved several modifications to generate a unique local specific immunity. Such modifications can be seen in the mechanisms of antigen processing, innate or acquired immune functions, the presence of M cells, and the segregation of the GALT from other mucosa associated lymphoid tissue (MALT) [11, 25, 36, 86, 87].
The mucosal innate immunity has two components, nonimmune and immune. The non-immune components include physico-chemical barriers including digestive enzymes, mucin, peristalsis, indigenous microbial flora, and the epithelial barrier with TJs [61] (Figure 6). The immune components include cellular and soluble elements. Nearly all classes of cells participate in natural immunity, including phagocytes, mast cells, IECs, and natural killer T cells (NK-T) [1, 2, 35]. Based upon the type of signals received from surface receptors of the IM, these cells phagocytize a microbe or antigen, secrete substances facilitating removal of the offending antigen (e.g., cryptidins), and recruit other cell types to produce pro-inflammatory substances (e.g., IL-8 from IECs and interferon-γ [IFN-γ] from NK-T cells) [36, 40]. These cells also remove altered (by injury or infection) host cells (e.g., granzymes and perforins from NK-cells) or modify specific immune responses (e.g., IL-4 from NK-T cells). Several of these responses are initiated by interactions between MAMPs and PRRs [40].
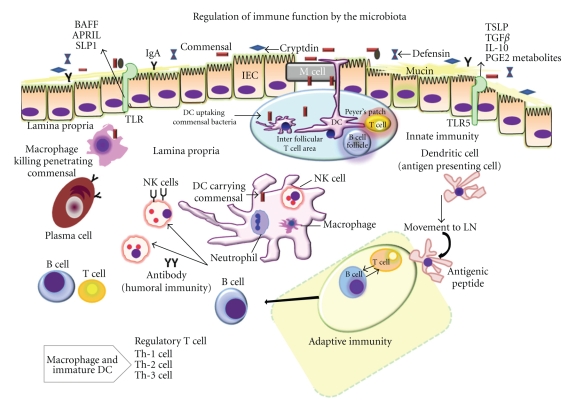
Figure 6.
Influenced by the microbiota, intestinal epithelial cells (IECs) elaborate cytokines, including thymic stromal lymphoprotein (TSLP), transforming growth factor (TGF), and interleukin-10 (IL-10), that can influence pro-inflammatory cytokine production by dendritic cells (DC) and macrophages present in the lamina propria (GALT) and Peyer's patches. Signals from commensal organisms may influence tissue-specific functions, resulting in T-cell expansion and regulation of the numbers of Th-1, Th-2, and Th-3 cells. Also modulated by the microbiota, other IEC derived factors, including APRIL (a proliferation-inducing ligand), B-cell activating factor (BAFF), secretory leucocyte peptidase inhibitor (SLPI), prostaglandin E2 (PGE2), and other metabolites, directly regulate functions of both antigen presenting cells and lymphocytes in the intestinal ecosystem. NK: natural killer cell; LN: lymph node; DC: dendritic cells. (Modified from Sharma R, Young C, Mshvildadze M, Neu J. Intestinal Microbiota Does It Play a Role in Diseases of the Neonate? NeoReviews Vol.10 No.4 2009 e166, with permission).
The receptors of the innate immune system are expressed on IECs and antigen presenting cells including monocytes, macrophages, B-cells, and dendritic-cells (DCs), [1, 85] (Figure 6). DCs in the lamina propria are specialized to regulate T cell immunity. Normally traversing through non-lymphoid tissue in immature form, DCs switch to immune-stimulatory mode upon encountering inflammatory stimuli. This process, referred to as maturation, changes the phenotype and function of DCs, including up-regulation of costimulatory and adhesion molecules and expression of distinct chemokine receptors [85, 86, 88, 89].
With respect to adaptive (acquired) immunity, helper T (Th) cells can be divided into four distinct cell types. Th1 cells secrete IL-2 and IFN-γ, a profile that supports the early events of T-cell and B-cell development (sIgG1 production) as well as cell mediated immunity (delayed-type hypersensitivity) [14, 34, 87, 88, 90–93]. Th2 cells, which predominantly secrete IL-4, IL-5, IL-6, IL-10, and IL-13, have a profile associated with humoral immunity (IgG4 and IgE production). Th3 cells, which predominantly secrete TGFβ, display a profile associated with suppression [88]. And finally, T-regulatory (T-reg1) cells, which predominantly secrete IL-10, also have a profile associated with suppression. Based on these phenotypes, T cells are either considered effector (Th1, Th2) or regulator (Th3, Treg1) T cells [40, 90]. The cell designated antigen (CD4+) cells which express CD25, the IL-2 receptor α chain (CD4 + CD25 + cells), may be particularly important T-regulatory cells capable of secreting IL-10 and TGF-β. These cytokines are responsible for diminishing the pro-inflammatory process and seem to prevent autoimmune gastritis and inflammatory bowel disease [86–90]. Through this regulatory mechanism, commensal organisms invoke tolerance (hyporesponsiveness) to self. Th0 cells exhibit a secretory pattern of cytokines that includes those associated with Th1 and Th2 patterns, and also represent newly stimulated T cells. Th0 cells are driven to become either Th1 cells by IL-12, which stimulates the transcription factor T-β or Th2 cells by IL-4, which stimulates the transcription factor GATA-3 [90–92].
Regulatory Treg and Th17 cells are two recently described subsets with opposing actions [90]. At a steady state, the lamina propria maintains a balanced population of cells of CD4 lineage, TH17 cells, and Treg cells. Th17 cells induce production of IL-17, a potent inflammatory regulator [88–93]. Production of IL-17 requires the presence of IL-6 and TGFβ activation, as well as a microbial trigger [93]. It has been suggested that a balance between Th17 and Treg is regulated through microbiota that can influence tolerance (hypo-responsiveness) or inflammatory response [92, 93]. Evidence is accumulating that targeting IL-17 signaling might prove useful in treating a variety of inflammatory diseases [88–93].
Chemokines present another important family of mediators involved in the immune-inflammatory response, activating chemo-attraction and leucocytes [2]. The receptors for chemokines (C, CC, CXC, CX3C) are seven membrane spanning G-protein-linked receptors that are coupled to cell activation through calcium mobilization and respond to cytokines of the CXC chemokine family. Interleukin-8 (IL-8 or CXL-8), induced by interferon gamma (Mig or CXCL9), interferon-inducible protein-10 (IP-10 or CXL10), and Interferon-inducible T cell chemo-attractant (I-TAC or CXCL11), belong to the CXC family that are expressed by enterocytes, presenting another mechanism of immune-inflammatory response by enterocytes [1, 11, 87, 94].
5. Conclusions
External to the IM, the microbiota, mucin, and antibacterial products (such as defensins and immunoglobulins) protect the host against pathogens. The layer of epithelial cells with several components of innate immunity including PRRs on the surface, NOD2/CARD15 intracellularly, and the paracelluar space sealed with TJA [40] present a relatively impermeable brush border. Then, important interactions take place under the control of the adaptive immune system in the lamina propria. The dynamic landscape of the IM in this manner is constantly adjusting to changing conditions while providing nutrition and also maintaining efficient immunity. In this respect, the symbiotic relationship of the IM with the microbiota is critical [3, 26, 40].
Manipulations of the microbiota to enhance its beneficial components that complement the human immune system with barrier-fortification are areas of active research [4, 23, 26, 34]. Disruption in the establishment of a stable microbiota (dysbiosis) is associated with diseases including inflammatory bowel diseases (IBD), obesity, atopy, and neonatal necrotizing enterocolitis (NEC) [44, 47, 70, 73]. The immature immune system and frequent exposure to antibiotics in neonatal intensive care units render premature infants, who are particularly susceptible to dysbiosis, more likely to develop NEC. Available conventional therapies for inflammatory intestinal diseases including NEC primarily target the infectious and inflammatory components of the diseases [3, 61, 73, 84]. The contribution of microbial communities in maintaining the integrity of the IM and in facilitating repair emphasizes the rationale for therapeutic exploitation of commensal organisms. There is evidence supporting a therapeutic role for probiotic strategies in treating certain diseases (e.g., IBD, NEC, diarrhea), but scientific validations and further research investigating the efficacy of such strategies without long-term immunological complications are warranted, particularly when such strategies apply to immune-incompetent hosts such as premature infants or cancer patients [5, 73, 76, 82, 86, 93, 94]. Understanding microbial-mucosal signaling components of inflammatory pathways and the mechanisms of how commensal organisms regulate these pathways should also provide new directions in treating and preventing these diseases.
Acknowledgment
This work was supported by the National Institutes of Health Grant RO1 HD05914301.
Abbreviations
- IM:
intestinal mucosa
- PRRs:
pattern recognition receptors
- MAMPs:
microbial-associated molecular patterns
- LPS:
lipopolysaccharide
- TLR:
Toll-like receptor
- NOD:
nucleotide-binding oligomerization domain
- NLRs:
NOD-like receptors
- IFN1:
interferon 1
- FPRs:
formylated peptide receptors
- NADPH:
nicotinamide adenine dinucleotide phosphate
- NOX:
NADPH oxide
- IECs:
intestinal epithelial cells
- NFκB:
nuclear factor kappa B
- Rel:
proteins coded by rel oncogenes
- MAPK:
mitogen-activated protein kinase
- IκB:
Inhibitory kappa B
- PPAR:
peroxisome proliferator-activated receptor
- TGF:
transforming growth factor-β
- Myd88:
myeloid differentiation primary-response gene 88
- FADD:
fast activated death domain
- IL:
interleukin
- IPAF:
intra-cellular IL-1β-converting enzyme protease-activator factor
- NAIP:
neuronal apoptosis inhibitory protein
- GALT:
gut-associated lymphoid tissue
- IEL:
intra-epithelial lymphocytes
- TCR:
T-cell receptor
- Ang4:
Angiogenein-4
- CARD:
caspase recruitment domain
- IκK:
inhibitory κB kinase
- TIR:
Toll-interleukin 1 receptor
- IRAK:
IL-1-R associated protein kinase
- TOLLIP:
Toll-interacting protein
- SIGIRR:
single immunoglobulin IL-1R-related protein
- SCFA:
short chain fatty acids
- CD14:
cell designated protein 14
- LBP:
lipopolysaccharide binding protein
- Ras:
reticulo endothelial-activating system
- MEK1/2:
Meiosis specific serine/threonine protein kinase
- Erk1/2:
extracellular signal-regulated kinase
- ADAM10:
adhesion and protease domain metallopeptidase 10
- GPCR:
G protein-coupled receptor
- Src:
rous sarcoma virus-cytoplasmic protein
- LTA:
lipoteichoic acid
- PAF:
platelet-activating factor
- EGF:
epidermal growth factor
- RELM-β:
resistin-like molecule
- TFF:
trefoil factors
- TJ:
tight junction
- AJ:
adherens junction
- ZO:
zona occludens
- JAMs:
junctional adhesion molecules
- CAR:
coxcackie virus and adenovirus receptor protein
- PI3K:
phosphatidylinositol 3-kinase
- NSP4:
nonstructural protein 4
- IBD:
inflammatory bowel disease
- MHC:
major histocompatibility complex
- RegIIIγ:
regenerating islet-derived 3 gamma
- EEC:
enteroendocrine
- TNF-α:
tumor necrosis factor α
- STa:
heat stable enterotoxin
- GC:
guanylate cyclase
- cGMP:
cyclic guanosine monophosphate
- MMP-7:
matrix metalloproteinase-7
- M-cells:
microfold-cells
- MALT:
mucosa associated lymphoid tissue
- NK-T:
natural killer T cells
- DCs:
dendritic cells
- Th2:
T helper 2 cells
- T-reg:
T regulatory
- TGF-β:
transforming growth factor β
- CXC:
chemokine family
- IP:
interferon inducible protein
- TAC:
T cell chemo-attractant.
References
- 1.McCracken VJ, Lorenz RG. The gastrointestinal ecosystem: a precarious alliance among epithelium, immunity and microbiota. Cellular Microbiology. 2001;3(1):1–11. doi: 10.1046/j.1462-5822.2001.00090.x. [DOI] [PubMed] [Google Scholar]
- 2.MacDonald TT, Gordon JN. Bacterial regulation of intestinal immune responses. Gastroenterology Clinics of North America. 2005;34(3):401–412. doi: 10.1016/j.gtc.2005.05.012. [DOI] [PubMed] [Google Scholar]
- 3.Neish AS. Microbes in gastrointestinal health and disease. Gastroenterology. 2009;136(1):65–80. doi: 10.1053/j.gastro.2008.10.080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hooper LV. Bacterial contributions to mammalian gut development. Trends in Microbiology. 2004;12(3):129–134. doi: 10.1016/j.tim.2004.01.001. [DOI] [PubMed] [Google Scholar]
- 5.Neu J. Perinatal and neonatal manipulation of the intestinal microbiome: a note of caution. Nutrition Reviews. 2007;65(6):282–285. doi: 10.1301/nr.2007.jun.282-285. [DOI] [PubMed] [Google Scholar]
- 6.Eckburg PB, Bik EM, Bernstein CN, et al. Diversity of the human intestinal microbial flora. Science. 2005;308(5728):1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Turnbaugh PJ, Ley RE, Hamady M, Fraser-Liggett CM, Knight R, Gordon JI. The human microbiome project. Nature. 2007;449(7164):804–810. doi: 10.1038/nature06244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Adlerberth I. Factors influencing the establishment of the intestinal microbiota in infancy. Nestle Nutrition Workshop Series. 2008;62:13–29. doi: 10.1159/000146245. [DOI] [PubMed] [Google Scholar]
- 9.Camp JG, Kanther M, Semova I, Rawls JF. Patterns and scales in gastrointestinal microbial ecology. Gastroenterology. 2009;136(6):1989–2002. doi: 10.1053/j.gastro.2009.02.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wick MJ, Madara JL, Fields BN, Normark SJ. Molecular cross talk between epithelial cells and pathogenic microorganisms. Cell. 1991;67(4):651–659. doi: 10.1016/0092-8674(91)90061-3. [DOI] [PubMed] [Google Scholar]
- 11.Akira S. Pathogen recognition by innate immunity and its signaling. Proceedings of the Japan Academy, Series B. 2009;85(4):143–156. doi: 10.2183/pjab.85.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Figdor CG, van Kooyk Y, Adema GJ. C-type lectin receptors on dendritic cells and langerhans cells. Nature Reviews Immunology. 2002;2(2):77–84. doi: 10.1038/nri723. [DOI] [PubMed] [Google Scholar]
- 13.Hooper LV, Wong MH, Thelin A, Hansson L, Falk PG, Gordon JI. Molecular analysis of commensal host-microbial relationships in the intestine. Science. 2001;291(5505):881–884. doi: 10.1126/science.291.5505.881. [DOI] [PubMed] [Google Scholar]
- 14.Thompson AJV, Locarnini SA. Toll-like receptors, RIG-I-like RNA helicases and the antiviral innate immune response. Immunology and Cell Biology. 2007;85(6):435–445. doi: 10.1038/sj.icb.7100100. [DOI] [PubMed] [Google Scholar]
- 15.Mai V, Morris JG., Jr. Colonic bacterial flora: changing understandings in the molecular age. Journal of Nutrition. 2004;134(2):459–464. doi: 10.1093/jn/134.2.459. [DOI] [PubMed] [Google Scholar]
- 16.Alverdy JC, Chang EB. The re-emerging role of the intestinal microflora in critical illness and inflammation: why the gut hypothesis of sepsis syndrome will not go away. Journal of Leukocyte Biology. 2008;83(3):461–466. doi: 10.1189/jlb.0607372. [DOI] [PubMed] [Google Scholar]
- 17.Migeotte I, Communi D, Parmentier M. Formyl peptide receptors: a promiscuous subfamily of G protein-coupled receptors controlling immune responses. Cytokine and Growth Factor Reviews. 2006;17(6):501–519. doi: 10.1016/j.cytogfr.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 18.Babbin BA, Jesaitis AJ, Ivanov AI, et al. Formyl peptide receptor-1 activation enhances intestinal epithelial cell restitution through phosphatidylinositol 3-kinase-dependent activation of Rac1 and Cdc42. Journal of Immunology. 2007;179(12):8112–8121. doi: 10.4049/jimmunol.179.12.8112. [DOI] [PubMed] [Google Scholar]
- 19.Bouskra D, Brezillon C, Berard M, et al. Lymphoid tissue genesis induced by commensals through NOD1 regulates intestinal homeostasis. Nature. 2008;456(7221):507–510. doi: 10.1038/nature07450. [DOI] [PubMed] [Google Scholar]
- 20.Abreu MT, Fukata M, Arditi M. TLR signaling in the gut in health and disease. Journal of Immunology. 2005;174(8):4453–4460. doi: 10.4049/jimmunol.174.8.4453. [DOI] [PubMed] [Google Scholar]
- 21.Neish AS. TLRS in the gut. II. Flagellin-induced inflammation and antiapoptosis. American Journal of Physiology. 2007;292(2):G462–G466. doi: 10.1152/ajpgi.00274.2006. [DOI] [PubMed] [Google Scholar]
- 22.Caicedo RA, Schanler RJ, Li N, Neu J. The developing intestinal ecosystem: implications for the neonate. Pediatric Research. 2005;58(4):625–628. doi: 10.1203/01.PDR.0000180533.09295.84. [DOI] [PubMed] [Google Scholar]
- 23.O’Hara AM, Shanahan F. Gut microbiota: mining for therapeutic potential. Clinical Gastroenterology and Hepatology. 2007;5(3):274–284. doi: 10.1016/j.cgh.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 24.Leaphart CL, Tepas JJ., III The gut is a motor of organ system dysfunction. Surgery. 2007;141(5):563–569. doi: 10.1016/j.surg.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 25.Marchesi J, Shanahan F. The normal intestinal microbiota. Current Opinion in Infectious Diseases. 2007;20(5):508–513. doi: 10.1097/QCO.0b013e3282a56a99. [DOI] [PubMed] [Google Scholar]
- 26.O’Hara AM, Shanahan F. The gut flora as a forgotten organ. EMBO Reports. 2006;7(7):688–693. doi: 10.1038/sj.embor.7400731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Round JL, Mazmanian SK. The gut microbiota shapes intestinal immune responses during health and disease. Nature Reviews Immunology. 2009;9(5):313–323. doi: 10.1038/nri2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rakoff-Nahoum S, Paglino J, Eslami-Varzaneh F, Edberg S, Medzhitov R. Recognition of commensal microflora by toll-like receptors is required for intestinal homeostasis. Cell. 2004;118(2):229–241. doi: 10.1016/j.cell.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 29.Mshvildadze M, Neu J, Mai V. Intestinal microbiota development in the premature neonate: establishment of a lasting commensal relationship? Nutrition Reviews. 2008;66(11):658–663. doi: 10.1111/j.1753-4887.2008.00119.x. [DOI] [PubMed] [Google Scholar]
- 30.Huse SM, Dethlefsen L, Huber JA, Welch DM, Relman DA, Sogin ML. Exploring microbial diversity and taxonomy using SSU rRNA hypervariable tag sequencing. PLoS Genetics. 2008;4(11, article e1000255) doi: 10.1371/journal.pgen.1000255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.DeSantis TZ, Brodie EL, Moberg JP, Zubieta IX, Piceno YM, Andersen GL. High-density universal 16S rRNA microarray analysis reveals broader diversity than typical clone library when sampling the environment. Microbial Ecology. 2007;53(3):371–383. doi: 10.1007/s00248-006-9134-9. [DOI] [PubMed] [Google Scholar]
- 32.Loy A, Lehner A, Lee N, et al. Oligonucleotide microarray for 16S rRNA gene-based detection of all recognized lineages of sulfate-reducing prokaryotes in the environment. Applied and Environmental Microbiology. 2002;68(10):5064–5081. doi: 10.1128/AEM.68.10.5064-5081.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Stappenbeck TS, Hooper LV, Gordon JI. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proceedings of the National Academy of Sciences of the United States of America. 2002;99(24):15451–15455. doi: 10.1073/pnas.202604299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Conroy M, Walker W. Intestinal immune health. Nestle Nutrition Workshop Series. 2008;62:111–121. doi: 10.1159/000146255. [DOI] [PubMed] [Google Scholar]
- 35.Ismail AS, Hooper LV. Epithelial cells and their neighbors. IV. Bacterial contributions to intestinal epithelial barrier integrity. American Journal of Physiology. 2005;289(5):G779–G784. doi: 10.1152/ajpgi.00203.2005. [DOI] [PubMed] [Google Scholar]
- 36.Macpherson AJ, Harris NL. Interactions between commensal intestinal bacteria and the immune system. Nature Reviews Immunology. 2004;4(6):478–485. doi: 10.1038/nri1373. [DOI] [PubMed] [Google Scholar]
- 37.Falk PG, Hooper LV, Midtvedt T, Gordon JI. Creating and maintaining the gastrointestinal ecosystem: what we know and need to know from gnotobiology. Microbiology and Molecular Biology Reviews. 1998;62(4):1157–1170. doi: 10.1128/mmbr.62.4.1157-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abrams GD, Bauer H, Sprinz H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab Investigation. 1963;12:355–364. [PubMed] [Google Scholar]
- 39.Bry L, Falk PG, Midtvedt T, Gordon JI. A model of host-microbial interactions in an open mammalian ecosystem. Science. 1996;273(5280):1380–1383. doi: 10.1126/science.273.5280.1380. [DOI] [PubMed] [Google Scholar]
- 40.Mukherjee S, Vaishnava S, Hooper LV. Multi-layered regulation of intestinal antimicrobial defense. Cellular and Molecular Life Sciences. 2008;65(19):3019–3027. doi: 10.1007/s00018-008-8182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Umesaki Y, Setoyama H, Matsumoto S, Okada Y. Expansion of alpha beta T-cell receptor-bearing intestinal intraepithelial lymphocytes after microbial colonization in germ-free mice and its independence from thymus. Immunology. 1993;79(1):32–37. [PMC free article] [PubMed] [Google Scholar]
- 42.Lefrancois L, Goodman T. In vivo modulation of cytolytic activity and Thy-1 expression in TCR-gamma delta+ intraepithelial lymphocytes. Science. 1989;243(4899):1716–1718. doi: 10.1126/science.2564701. [DOI] [PubMed] [Google Scholar]
- 43.Umesaki Y, Setoyama H, Matsumoto S, Imaoka A, Itoh K. Differential roles of segmented filamentous bacteria and clostridia in development of the intestinal immune system. Infection and Immunity. 1999;67(7):3504–3511. doi: 10.1128/iai.67.7.3504-3511.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Biol MC. Nutritional and developmental variations of intestinal glycosylation. Nutrition. 1992;8(5):368–369. [PubMed] [Google Scholar]
- 45.Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nature Immunology. 2003;4(3):269–273. doi: 10.1038/ni888. [DOI] [PubMed] [Google Scholar]
- 46.Umesaki Y, Okada Y, Matsumoto S, Imaoka A, Setoyama H. Segmented filamentous bacteria are indigenous intestinal bacteria that activate intraepithelial lymphocytes and induce MHC class II molecules and fucosyl asialo GM1 glycolipids on the small intestinal epithelial cells in the ex-germ-free mouse. Microbiology and Immunology. 1995;39(8):555–562. doi: 10.1111/j.1348-0421.1995.tb02242.x. [DOI] [PubMed] [Google Scholar]
- 47.Kelly D, Conway S, Aminov R. Commensal gut bacteria: mechanisms of immune modulation. Trends in Immunology. 2005;26(6):326–333. doi: 10.1016/j.it.2005.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Li N, Russell WM, Douglas-Escobar M, Hauser N, Lopez M, Neu J. Live and heat-killed Lactobacillus rhamnosus GG: effects on proinflammatory and anti-inflammatory cytokines/chemokines in gastrostomy-fed infant rats. Pediatric Research. 2009;66(2):203–207. doi: 10.1203/PDR.0b013e3181aabd4f. [DOI] [PubMed] [Google Scholar]
- 49.Yan F, Cao H, Cover TL, Whitehead R, Washington MK, Polk DB. Soluble proteins produced by probiotic bacteria regulate intestinal epithelial cell survival and growth. Gastroenterology. 2007;132(2):562–575. doi: 10.1053/j.gastro.2006.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Karin M, Ben-Neriah Y. Phosphorylation meets ubiquitination: the control of NF-κB activity. Annual Review of Immunology. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 51.Neish AS, Gewirtz AT, Zeng H, et al. Prokaryotic regulation of epithelial responses by inhibition of IκB-α ubiquitination. Science. 2000;289(5484):1560–1563. doi: 10.1126/science.289.5484.1560. [DOI] [PubMed] [Google Scholar]
- 52.Dharmani P, Srivastava V, Kissoon-Singh V, Chadee K. Role of intestinal mucins in innate host defense mechanisms against pathogens. Journal of Innate Immunity. 2009;1(2):123–135. doi: 10.1159/000163037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johansson ME, Phillipson M, Petersson J, Velcich A, Holm L, Hansson GC. The inner of the two Muc2 mucin-dependent mucus layers in colon is devoid of bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(39):15064–15069. doi: 10.1073/pnas.0803124105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yan Y, Shirakabe K, Werb Z. The metalloprotease Kuzbanian (ADAM10) mediates the transactivation of EGF receptor by G protein-coupled receptors. Journal of Cell Biology. 2002;158(2):221–226. doi: 10.1083/jcb.200112026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. American Journal of Clinical Nutrition. 2001;73(6):1131S–1141S. doi: 10.1093/ajcn/73.6.1131S. [DOI] [PubMed] [Google Scholar]
- 56.Linden SK, Florin THJ, McGuckin MA. Mucin dynamics in intestinal bacterial infection. PLoS ONE. 2008;3(12, article e3952) doi: 10.1371/journal.pone.0003952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Suemori S, Lynch-Devaney K, Podolsky DK. Identification and characterization of rat intestinal trefoil factor: tissue- and cell-specific member of the trefoil protein family. Proceedings of the National Academy of Sciences of the United States of America. 1991;88(24):11017–11021. doi: 10.1073/pnas.88.24.11017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Podolsky DK. Mucosal immunity and inflammation V. Innate mechanisms of mucosal defense and repair: the best offense is a good defense. American Journal of Physiology. 1999;277(3):G495–G499. doi: 10.1152/ajpgi.1999.277.3.G495. [DOI] [PubMed] [Google Scholar]
- 59.Artis D, Wang ML, Keilbaugh SA, et al. RELMβ/FIZZ2 is a goblet cell-specific immune-effector molecule in the gastrointestinal tract. Proceedings of the National Academy of Sciences of the United States of America. 2004;101(37):13596–13600. doi: 10.1073/pnas.0404034101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Krimi RB, Kotelevets L, Dubuquoy L, et al. Resistin-like molecule β regulates intestinal mucous secretion and curtails TNBS-induced colitis in mice. Inflammatory Bowel Diseases. 2008;14(7):931–941. doi: 10.1002/ibd.20420. [DOI] [PubMed] [Google Scholar]
- 61.Liu Z, Li N, Neu J. Tight junctions, leaky intestines, and pediatric diseases. Acta Paediatrica. 2005;94(4):386–393. doi: 10.1111/j.1651-2227.2005.tb01904.x. [DOI] [PubMed] [Google Scholar]
- 62.Ivanov AI, Nusrat A, Parkos CA. Endocytosis of epithelial apical junctional proteins by a clathrin-mediated pathway into a unique storage compartment. Molecular Biology of the Cell. 2004;15(1):176–188. doi: 10.1091/mbc.E03-05-0319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Li N, Neu J. Glutamine deprivation alters intestinal tight junctions via a PI3-K/Akt mediated pathway in caco-2 cells. Journal of Nutrition. 2009;139(4):710–714. doi: 10.3945/jn.108.101485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hossain Z, Hirata T. Molecular mechanism of intestinal permeability: interaction at tight junctions. Molecular BioSystems. 2008;4(12):1181–1185. doi: 10.1039/b800402a. [DOI] [PubMed] [Google Scholar]
- 65.Wittchen ES, Haskins J, Stevenson BR. Protein interactions at the tight junction. Actin has multiple binding partners, and ZO-1 forms independent complexes with ZO-2 and ZO-3. Journal of Biological Chemistry. 1999;274(49):35179–35185. doi: 10.1074/jbc.274.49.35179. [DOI] [PubMed] [Google Scholar]
- 66.Cani PD, Possemiers S, Van de Wiele T, et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut. 2009;58(8):1091–1103. doi: 10.1136/gut.2008.165886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Prasad S, Mingrino R, Kaukinen K, et al. Inflammatory processes have differential effects on claudins 2, 3 and 4 in colonic epithelial cells. Laboratory Investigation. 2005;85(9):1139–1162. doi: 10.1038/labinvest.3700316. [DOI] [PubMed] [Google Scholar]
- 68.Tafazoli F, Zeng CQ, Estes MK, Magnusson K-E, Svensson L. NSP4 enterotoxin of rotavirus induces paracellular leakage in polarized epithelial cells. Journal of Virology. 2001;75(3):1540–1546. doi: 10.1128/JVI.75.3.1540-1546.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Meddings J. The significance of the gut barrier in disease. Gut. 2008;57(4):438–440. doi: 10.1136/gut.2007.143172. [DOI] [PubMed] [Google Scholar]
- 70.Cash HL, Whitham CV, Behrendt CL, Hooper LV. Symbiotic bacteria direct expression of an intestinal bactericidal lectin. Science. 2006;313(5790):1126–1130. doi: 10.1126/science.1127119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salzman NH, Underwood MA, Bevins CL. Paneth cells, defensins, and the commensal microbiota: a hypothesis on intimate interplay at the intestinal mucosa. Seminars in Immunology. 2007;19(2):70–83. doi: 10.1016/j.smim.2007.04.002. [DOI] [PubMed] [Google Scholar]
- 72.Vaishnava S, Behrendt CL, Ismail AS, Eckmann L, Hooper LV. Paneth cells directly sense gut commensals and maintain homeostasis at the intestinal host-microbial interface. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(52):20858–20863. doi: 10.1073/pnas.0808723105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sonnenburg JL, Chen CT, Gordon JI. Genomic and metabolic studies of the impact of probiotics on a model gut symbiont and host. PLoS Biology. 2006;4(12, article e413) doi: 10.1371/journal.pbio.0040413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Roth KA, Hertz JM, Gordon JI. Mapping enteroendocrine cell populations in transgenic mice reveals an unexpected degree of complexity in cellular differentiation within the gastrointestinal tract. Journal of Cell Biology. 1990;110(5):1791–1801. doi: 10.1083/jcb.110.5.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lin PW, Simon PO, Jr., Gewirtz AT, et al. Paneth cell cryptdins act in vitro as apical paracrine regulators of the innate inflammatory response. Journal of Biological Chemistry. 2004;279(19):19902–19907. doi: 10.1074/jbc.M311821200. [DOI] [PubMed] [Google Scholar]
- 76.Samuel BS, Shaito A, Motoike T, et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(43):16767–16772. doi: 10.1073/pnas.0808567105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Hornef MW, Putsep K, Karlsson J, Refai E, Andersson M. Increased diversity of intestinal antimicrobial peptides by covalent dimer formation. Nature Immunology. 2004;5(8):836–843. doi: 10.1038/ni1094. [DOI] [PubMed] [Google Scholar]
- 78.Salzman NH, Ghosh D, Huttner KM, Paterson Y, Bevins CL. Protection against enteric salmonellosis in transgenic mice expressing a human intestinal defensin. Nature. 2003;422(6931):522–526. doi: 10.1038/nature01520. [DOI] [PubMed] [Google Scholar]
- 79.Froy O, Chapnik N, Miskin R. Mouse intestinal cryptdins exhibit circadian oscillation. FASEB Journal. 2005;19(13):1920–1922. doi: 10.1096/fj.05-4216fje. [DOI] [PubMed] [Google Scholar]
- 80.Lussier C, Basora N, Bouatrouss Y, Beaulieu J-F. Integrins as mediators of epithelial cell-matrix interactions in the human small intestinal mucosa. Microscopy Research and Technique. 2000;51(2):169–178. doi: 10.1002/1097-0029(20001015)51:2<169::AID-JEMT8>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 81.Saenz SA, Taylor BC, Artis D. Welcome to the neighborhood: epithelial cell-derived cytokines license innate and adaptive immune responses at mucosal sites. Immunological Reviews. 2008;226(1):172–190. doi: 10.1111/j.1600-065X.2008.00713.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Canny GO, McCormick BA. Bacteria in the intestine, helpful residents or enemies from within? Infection and Immunity. 2008;76(8):3360–3373. doi: 10.1128/IAI.00187-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Spahn TW, Muller MK, Domschke W, Kucharzik T. Role of lymphotoxins in the development of Peyer’s patches and mesenteric lymph nodes: relevance to intestinal inflammation and treatment. Annals of the New York Academy of Sciences. 2006;1072:187–193. doi: 10.1196/annals.1326.029. [DOI] [PubMed] [Google Scholar]
- 84.Neu J, Chen M, Beierle E. Intestinal innate immunity: how does it relate to the pathogenesis of necrotizing enterocolitis. Seminars in Pediatric Surgery. 2005;14(3):137–144. doi: 10.1053/j.sempedsurg.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 85.Niess JH, Reinecker H-C. Dendritic cells: the commanders-in-chief of mucosal immune defenses. Current Opinion in Gastroenterology. 2006;22(4):354–360. doi: 10.1097/01.mog.0000231807.03149.54. [DOI] [PubMed] [Google Scholar]
- 86.MacDonald TT, Monteleone G. Immunity, inflammation, and allergy in the gut. Science. 2005;307(5717):1920–1925. doi: 10.1126/science.1106442. [DOI] [PubMed] [Google Scholar]
- 87.Forchielli ML, Walker WA. The role of gut-associated lymphoid tissues and mucosal defence. British Journal of Nutrition. 2005;93(supplement 1):S41–S48. doi: 10.1079/bjn20041356. [DOI] [PubMed] [Google Scholar]
- 88.Eisenstein EM, Williams CB. The Treg/Th17 cell balance: a new paradigm for autoimmunity. Pediatric Research. 2009;65(supplement 5):26R–31R. doi: 10.1203/PDR.0b013e31819e76c7. [DOI] [PubMed] [Google Scholar]
- 89.Niessner M, Volk BA. Altered Th1/Th2 cytokine profiles in the intestinal mucosa of patients with inflammatory bowel disease as assessed by quantitative reversed transcribed polymerase chain reaction (RT-PCR) Clinical and Experimental Immunology. 1995;101(3):428–435. doi: 10.1111/j.1365-2249.1995.tb03130.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Ivanov II, Frutos RL, Manel N, et al. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host and Microbe. 2008;4(4):337–349. doi: 10.1016/j.chom.2008.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Andoh A, Yagi Y, Shioya M, Nishida A, Tsujikawa T, Fujiyama Y. Mucosal cytokine network in inflammatory bowel disease. World Journal of Gastroenterology. 2008;14(33):5154–5161. doi: 10.3748/wjg.14.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Honda K, Takeda K. Regulatory mechanisms of immune responses to intestinal bacteria. Mucosal Immunology. 2009;2(3):187–196. doi: 10.1038/mi.2009.8. [DOI] [PubMed] [Google Scholar]
- 93.Ivanov S, Linden A. Interleukin-17 as a drug target in human disease. Trends in Pharmacological Sciences. 2009;30(2):95–103. doi: 10.1016/j.tips.2008.11.004. [DOI] [PubMed] [Google Scholar]
- 94.Mowat AM. Anatomical basis of tolerance and immunity to intestinal antigens. Nature Reviews Immunology. 2003;3(4):331–341. doi: 10.1038/nri1057. [DOI] [PubMed] [Google Scholar]