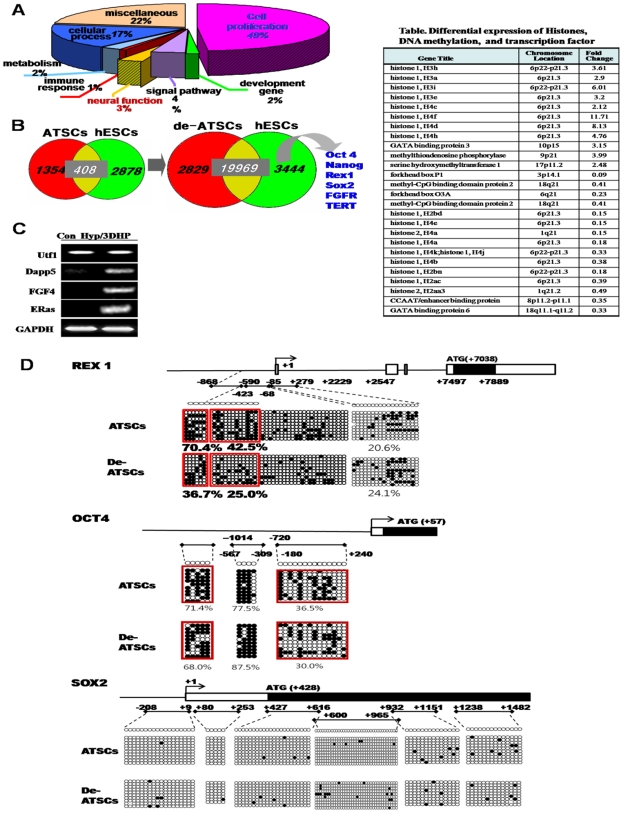
Figure 2. Functional categorization of differentially expressed gene profile and epigenetic reprogramming of stemness genes in de-ATSC.
(A) Functional categorization of genes in de-ATSC. (B) Commonly expressed genes between the de-ATSC and hESC. Also, many kinds of histones and DNA methylation-related transcription factors and enzymes are overexpressed after ATSC de-differentiation (Table). (C) Differential embryonic genes, Utf1, Dapp5, FGF4, and Eras expressions in de-ATSCs. (D) Evaluation of epigenetic modifications through methylation analysis on promoter regions through bisulfite modification and sequencing of genomic DNA. The related method was explained in Materials and Methods S1.