Figure 6.
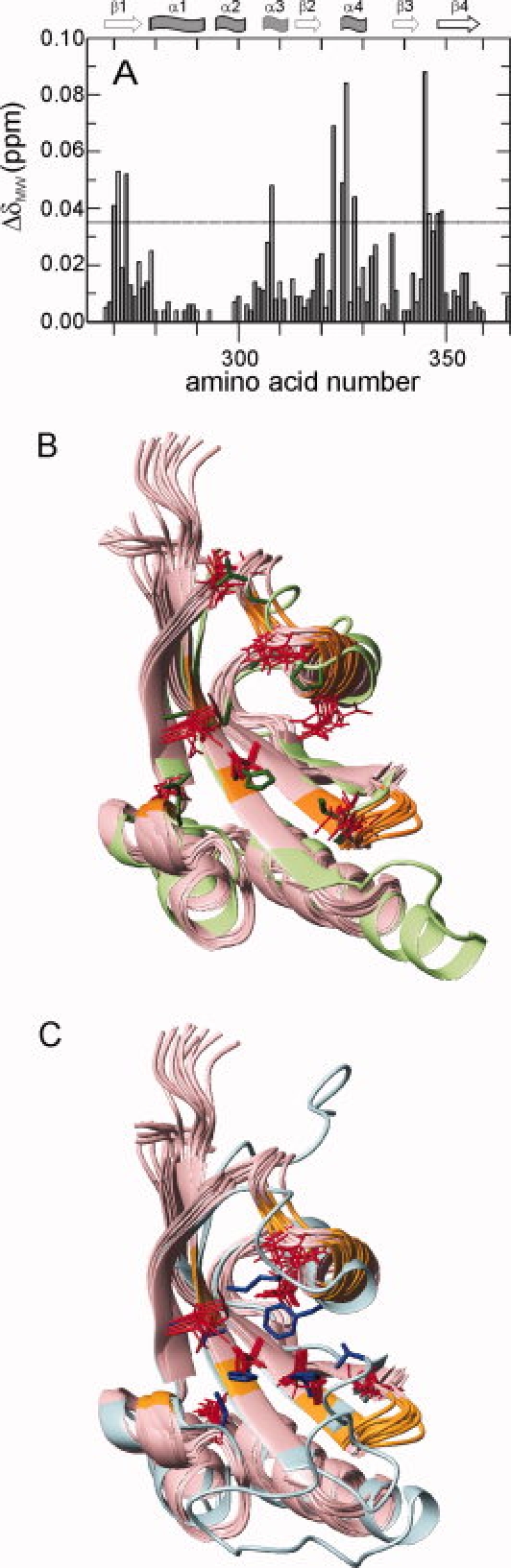
Peptide binding site of PpiD*. (A) The change in the averaged weighted chemical shift ΔδMW, caused by titration with Suc-Ala-Leu-Pro-Phe-pNA) at a ratio of 4:1 (2.29 mM peptide, 0.59 mM PpiD*) at 25°C, 0.1M K phosphate and pH 7.0 is plotted as a function of the amino acid sequence. Secondary structure elements are indicated on top. The threshold of 0.035 ppm is indicated by the horizontal line. The backbone of residues of PpiD* with a ΔδMW larger than this threshold (R270, Y271, I273, S308, I323, D325, E326, K328, S345, V346, F348, L349) are colored in orange in (B) and (C). (B) Overlay of the solution structure of PpiD* (in pink) and the crystal structure of the first parvulin domain of SurA (2pv2.pdb, in light green) in ribbon representation. The side chains of the active site residues of SurA (H178, Q223, M231, E238, L239, P240, V263) are shown in green, the corresponding side chains of PpiD* (I273, S308, M315, T322, P324, E326, V346) in red. (C) Overlay of the solution structure of PpiD* (in pink) and the crystal structure of human Pin1 (1pin.pdb, in cyan) in ribbon representation. The side chains of the active site residues of Pin1 (H59, C113, L121, M130, F134, T152, H157) are shown in blue, the corresponding side chains of PpiD* (I273, D305, M315, P324, L327, S344, L349) in red.
