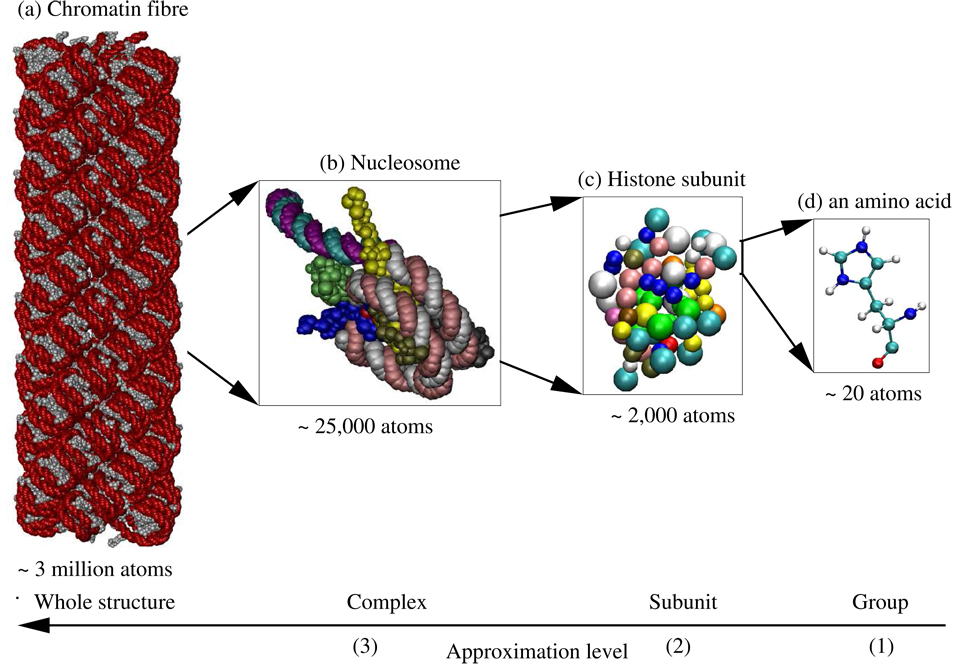
Figure 1.
Example of the natural partitioning of a chromatin fibre. (a) The fibre is made up of 100 nucleosome complexes. The individual nucleotide groups in the fibre are represented as red beads and amino acid groups as grey beads. (b) Each complex (level 3) is made up of 13 subunits with the segments of DNA linking nucleosome complexes being treated as separate subunits. A complex is shown here with each subunit represented in a different color. (c) Each subunit (level 2) is made up of 49–142 groups. The linker histone subunit is shown here with the groups colored by the type of amino acid. (d) Each group (level 1) is made up of 7–32 atoms (level 0). A histidine amino acid group is shown here with atoms represented as small spheres and covalent bonds between the atoms represented as links. The atoms are colored by the type of atom. The total fibre consists of approximately 3 million atoms. The images were rendered using VMD.68 For clarity only 10 of the 13 subunits are shown in (a) and (b).