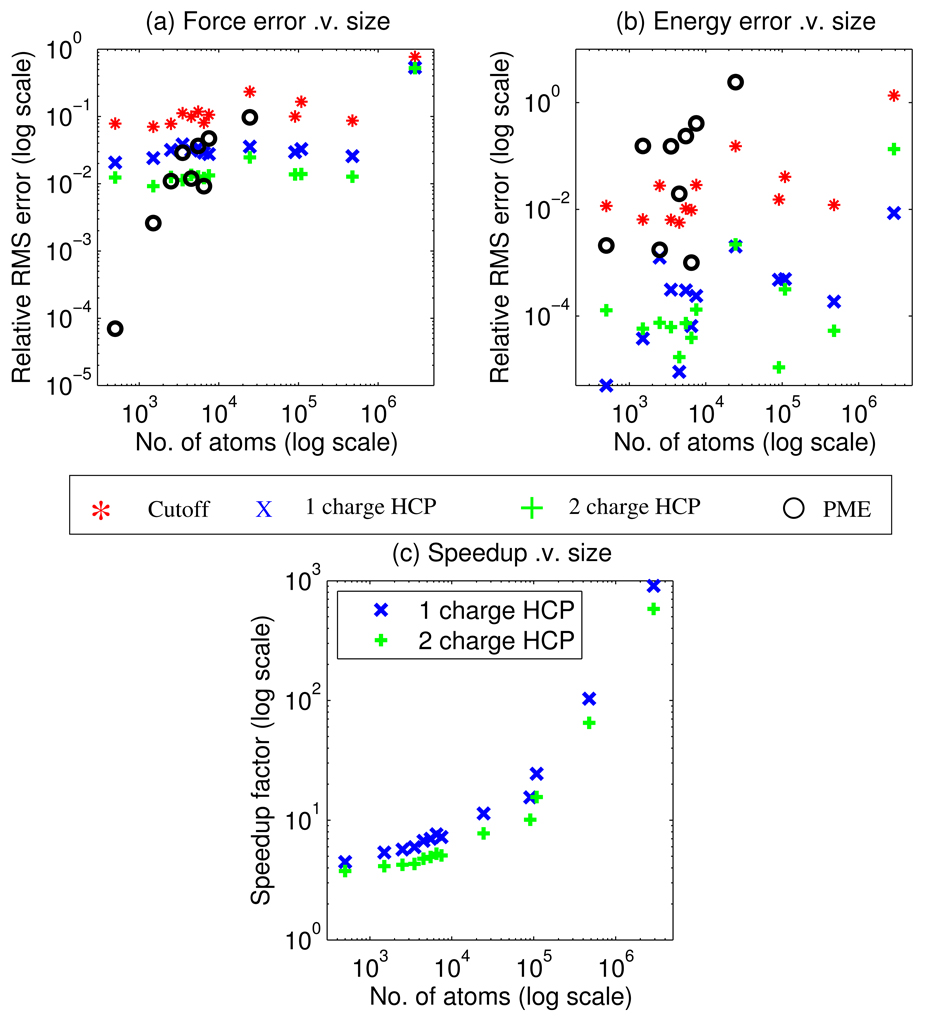
Figure 3.
Accuracy and speed-up comparison for a sample set of 600 biomolecular structures in vacuum. (a) Relative force error shows that for electrostatic force the hierarchical charge partitioning (HCP) is more accurate than the spherical cutoff method, and on average approaches the accuracy of the particle mesh Ewald (PME) method for all but very small structures. (b) Relative error in energy shows that for electrostatic energy the HCP is on average more accurate than both the spherical cutoff and PME methods. (c) Speedup factor compared to the exact all-atom calculation shows that the HCP can be multiple orders of magnitude faster than the exact O(N2) computation for large structures. The above computations use the following parameters. Spherical cutoff distance = 10 Å, the HCP threshold distances of h1 = 10 Å, h2 = 70 Å, and h3 = 125 Å, and PME cutoff distance = 10 Å. RMS error and speed-up factor for individual structures are grouped into bins, based on structure size in increments of 1000 atoms, and plotted at the midpoint of each bin.