Abstract
Purpose:
Chromatin modifying compounds that inhibit the activity of histone deacetylases have shown potency as radiosensitizers but the action of these drugs at a molecular level is not clear. Here we investigated the effect of SAHA on DNA breaks, their repair and induction of rearrangements.
Methods and Materials:
The effect of SAHA on both clonogenic survival and repair (SLDr) was assessed using SCC-25, MCF7 and TK6 cell lines. In order to study unique DNA double strand breaks, anti-CD95 antibody was employed that introduces a DNA double strand break at a known location within 11q23. The effect of SAHA on DNA cleavage and rearrangements were analyzed by ligation-mediated PCR and inverse PCR respectively.
Results:
SAHA acts as radiosensitizer at 1 μM with dose enhancement factors at 10% survival of: SCC-25 - 1.24±0.05, MCF7 - 1.16±0.09 and TK6 - 1.17±0.05 and reduced the capacity of SCC-25 cells to repair radiation induced lesions. Additionally, SAHA treatment diffused site-specific fragmentation over at least 1 kbp in TK6 cells. Chromosomal rearrangements produced in TK6 cells exposed to SAHA showed a reduction in microhomology at the breakpoint between 11q23 and partner chromosomes.
Conclusion:
SAHA shows efficacy as a radiosensitizer at clinically obtainable levels. In its presence, targeted DNA strand breaks occur over an expanded region indicating increased chromatin access. The rejoining of such breaks is degraded by SAHA when measured as either rearrangements at the molecular level and rejoining that contributes to cell survival.
Keywords: SAHA, Sub-lethal damage repair, DNA double strand breaks, DNA rearrangements, radiosensitizer
Introduction
The addition of chemotherapy to radiation treatment is a mainstay of current clinical practice with DNA damage being a major target of both (1). More recently, chromatin modifying compounds that inhibit the activity of histone deacetylases (HDACs) have been used in conjunction with irradiation (2). Several in vitro studies have supported the fact that targeting HDACs with HDAC inhibitors (HDACIs) can increase radiosensitivity (3-6). This observation has been attributed to the more open chromatin structure in the presence of HDACIs (7). Additionally, biochemical studies have attributed the radiosensitization effects of HDACIs to the interplay between HDACs and DNA damage response pathway proteins. ATM, the gene mutated in ataxia telangiectasia and linked to increased radiation sensitivity has been found to interact with HDAC1 (8). This gene has also been shown to be involved in histone acetylation-mediated gene regulation with the use of HDACIs (9). These various studies suggest that radiation sensitivity and radiation dependent DNA signaling can both be modified with HDACIs.
Several HDACIs have been developed and their action studied both in vivo and in vitro. These HDACIs can induce growth arrest, differentiation, and apoptosis in various cancer cell lines (10-12). Recently, suberoylanilide hydroxyamic acid (SAHA; vorinostat; Zolinza®), a broad inhibitor of class I and II of HDACs (13), has been approved by the FDA for treatment of cutaneous T-cell lymphoma (CTCL) (14). Subsequent single agent clinical studies of SAHA have shown that although it has limited activity against solid and hematological malignancies, the drug was well tolerated, thus had potential for clinical benefits as a part of combination therapy (15-17). In a preclinical study, pre-treatment with SAHA followed by irradiation showed enhanced toxicity in prostate and glioma cancer cell lines (3). Another study using leukemia cell lines showed that SAHA can enhance the effect of cytosine arabinoside and etoposide; two widely used anti-cancer drugs (18). Based on the success of in vitro studies, several clinical trials are being conducted at present to study the efficacy of SAHA in combination with radiation and chemotherapeutic drugs in malignancies of the breast, lung, brain and blood (19).
In order to understand the molecular mechanisms by which SAHA modulates the affect of DNA damaging agents, experiments were designed targeting the 11q23 region. This region has been shown to undergo active fragmentation upon exposure to various genotoxic agents, making it a suitable model to study the effect of SAHA on DNA damage and repair at the molecular level (20-22). In addition, we evaluated the radiosensitization potential of SAHA in cell lines representing three different systems, SCC-25 (head and neck carcinoma), MCF7 (breast cancer) and TK6 (lymphoblastoid).
Materials and Methods
Cell Lines
Cell lines used in this study were obtained from the American Type Culture Collection (ATCC, Manassas, VA). The TK6 cells were maintained in RPMI-1640 (ATCC) supplemented with 10% FBS. The MCF7 cells were maintained in AMEM (Gibco, Carlsbad, CA) containing 10% FBS, 0.01 mg/ml of bovine insulin, 1% penicillin-streptomycin, and 1% L-glutamine. The SCC-25 were maintained in a 1:1 mixture of DMEM (Cellgro, Manassas, VA) and Ham's F12 medium (Gibco) supplemented with 10% FBS, 1% penicillin-streptomycin, and 400 ng/ml hydrocortisone.
Clonogenic Cell Survival Assay
Exponentially growing MCF7 and SCC-25 cells were treated with either 0.5 μM or 1.0 μM of SAHA dissolved in DMSO for 24 h. The control group was treated with DMSO only. Cells were washed 3x with PBS and supplied with fresh medium prior to irradiation (IR). After IR, cells were harvested by trypsinization, counted, and seeded within a range of 100 to 16,000 cells per dish. After 14 d, cell colonies were fixed and stained with 1% crystal violet. Colonies of ≥ 50 cells were counted to determine the surviving fraction. Radiation survival curves were constructed by normalizing to the number of colonies surviving SAHA alone. The survival of TK6 cells after treatment was determined by a limiting dilution assay. Depending on the drug and IR dosages, cell concentrations ranged from 0.25 to128 cells/well. Cells were treated with DMSO or various concentrations of SAHA for 24 h. After drug treatment, cells were irradiated at 0, 2, 4, and 6 Gy. Following IR, plates were analyzed as described before (23).
Extraction of Cellular Histones
Cells were harvested after SAHA treatment, washed 2x with ice-cold 1X PBS and centrifuged at 13,000 rpm for 5 min at 4°C. Cells were suspended in Triton Extraction Buffer [TEB: PBS containing 0.5% Triton X-100 (v/v), 2 mM PMSF, 0.02% (v/v) NaN3] at a cell density of 106 cells/100 μl, lysed on ice for 10 min, centrifuged at 2000 rpm for 10 min and the pellet washed with TEB. Subsequently the pellet was suspended in 0.2 N HCl at a cell density of 106 cells/25 μl, and acid extraction was carried out overnight at 4°C, the suspension centrifuged for 10 min at 2000 rpm and the supernatant collected.
Detection of acetylated Histone H4 and caspase-3 proteins
Ten μg of protein from cells treated with SAHA was run on a polyacrylamide gel and transferred to a PVDF membrane. After transfer, the membrane was blocked for 1 h in 3% nonfat milk diluted in PBS and incubated overnight at 4°C with the primary antibody (anti-acetylated H4, Millipore, Billerica, MA) at 1:2000 dilution. After washing, the membrane was incubated for 1 h with the secondary antibody (Goat anti-rabbit-HRP, Pierce, Rockford, IL) at 1:5000 dilution and developed using SuperSignal West Dura (Thermo Scientific, Waltham, MA). For caspase-3 analysis total cellular proteins were extracted as described before (23). Cleavage of caspase-3 was detected by using anti-caspase-3 antibody (Cell Signaling Technology, Danvers, MA) that probes for both full length caspase-3 (35 kDa) and the large fragment of caspase-3 resulting from cleavage (17 kDa). The primary antibody was used at a dilution of 1:250. The secondary antibody (Goat anti-rabbit-HRP, Jackson ImmunoResearch, West Grove, PA) was used at a dilution of 1:5000.
Sub-Lethal Damage Repair Clonogenic Assay
SCC-25 cells were treated with either DMSO or 0.5 μM SAHA using cells at 75% confluence. After 24 h the cells were washed 3x in PBS and irradiated in fresh media. A time course was performed to measure the effect of recovery time between the first and second IR treatment. An initial dose of 2 Gy was administered to the cells. Recovery times from 0 to 120 min were used, followed by a second dose of 2 Gy. Subsequently, cells were washed 1x in PBS and trypsinized to yield a single cell suspension. Cells were counted and seeded within a range of 150 to 800 cells per dish and analyzed by clonogenic assay as above.
Ligation-mediated PCR
Ligation-mediated PCR (LM-PCR) was used to analyze the location and intensity of DNA fragmentation at 11q23. TK6 cells were incubated with SAHA for 24 h followed by treatment with anti-CD95 antibody (0.5 μg/ml; eBioscience, Inc., San Diego, CA) to induce DNA fragmentation. After 4 h of treatment, genomic DNA was collected and ligated with the linker. The details on making the linker, ligation to genomic DNA and subsequent PCR have been described before (21, 24). The PCR products from the LM-PCR reaction were transferred from to a nylon membrane. The hybridization was done with a biotinylated probe generated using NEBlot Phototope kit (New England Biolabs, Ipswich, MA) specific to the 11q23 region (20). The probe was detected using the Phototope – Star detection Kit (New England Biolabs).
Inverse PCR
TK6 cells were treated with DMSO or with 0.5 μM/2.0 μM of SAHA for 24 h, followed by treatment with anti-CD95 antibody for 4 h (0.5 μg/ml; eBioscience). The IPCR was conducted as described (24). The templates made from 11q23 ligated to partner chromosomes were amplified using a semi-nested PCR procedure. Each template was treated with PvuII restriction enzyme to limit amplification of normal, un-rearranged material. Aberrant bands were extracted, sequenced and analyzed by reference to the BLAST database at NCBI. To reduce the possibility of artifacts being mistaken for rearrangements, all rearrangements had templates with confirmed re-ligation at the appropriate Sau3A1 cleavage site. In addition, both final primer sites had to be present within the sequence analyzed. These precautions substantially reduce the possibilities of errors in interpretation (25).
Results
SAHA enhances radiosentivity
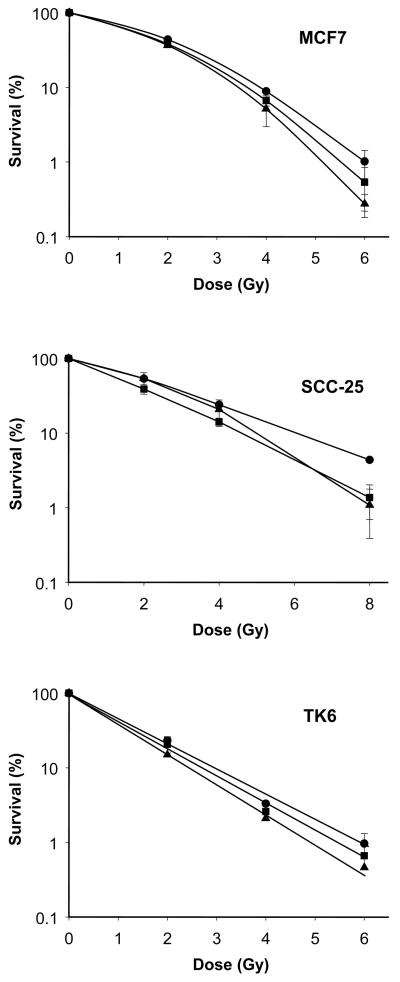
We were interested to determine if SAHA could act as a radiosensitizer at clinically obtainable levels of 0.5 μM and 1.0 μM using the clonogenic survival assay. SAHA pre-treatment decreased the survival fraction following irradiation for all three cell lines (Fig. 1). The dose enhancement factor (DEF) was calculated at 10% survival, Drads/Drads+drug, using the 1.0 μM SAHA survival curve. The DEF's were as follows: SCC-25 - 1.24±0.05, MCF7 - 1.16±0.09 and TK6 - 1.17±0.05. For the TK6 cell line only, clonogenic survivals were also conducted using a 2.0 μM SAHA pre-treatment, followed by irradiation. It was observed that the DEF at 10% survival increased to 1.34±0.17. Including the data from Fig. 1, a clear dose response was obtained with SAHA treatment in TK6 cell line, with DEF's of 1.09 + .05, 1.17+.05 and 1.34+.17 at 0.5, 1 and 2 μM respectively. For these experiments, average control and 0.5 μM SAHA treatment plating efficiencies were similar and varied between 17 and 30% for MCF7 and SCC-25 and 2-3% for TK6 cells. A slight reduction in plating efficiency was noted for all cell systems when 1.0 μM SAHA was used.
Fig. 1.
Clonogenic survival assay demonstrating the radiosensitizer capacity of SAHA. MCF7, SCC-25 and TK6 cell lines were treated with 0.0 μM (●), 0.5 μM (■) and 1.0 μM (▲) of SAHA for 24 h and then exposed to graded doses of ionizing radiation. Each point on the survival curve represents the surviving fraction ± SE from three separate experiments. Error bars are shown unless smaller than the symbol size.
SAHA affects the repair capacity of SCC-25
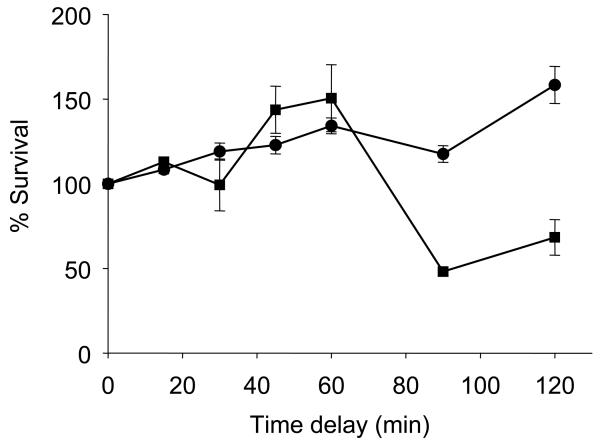
It was observed that the repair capacity of SAHA treated cells is similar to that of the control until the interval between the two irradiation doses exceeded 60 min. With a time interval of more than 60 min, the repair capacity of SAHA treated samples was significantly compromised (Fig. 2). These data indicate that SAHA can affect the repair capacity of SCC-25 at low concentrations and this could contribute to the ability of SAHA to act as a radiosensitizing agent.
Fig. 2.
Sub-lethal damage repair assay showing the compromised repair capacity of SCC-25 cells due to treatment with SAHA. SCC-25 cells were pre-treated with either 0.0 μM (●) or 0.5 μM (■) for 24 h and then two doses of 2 Gy each were delivered either simultaneously or at increasing times apart. SCC-25 cells showed decreased repair capacity when the interval between two radiation doses was more than 60 min. Each point on the survival curve represents the surviving fraction ± SE from three separate experiments.
SAHA treatment enhances histone acetylation
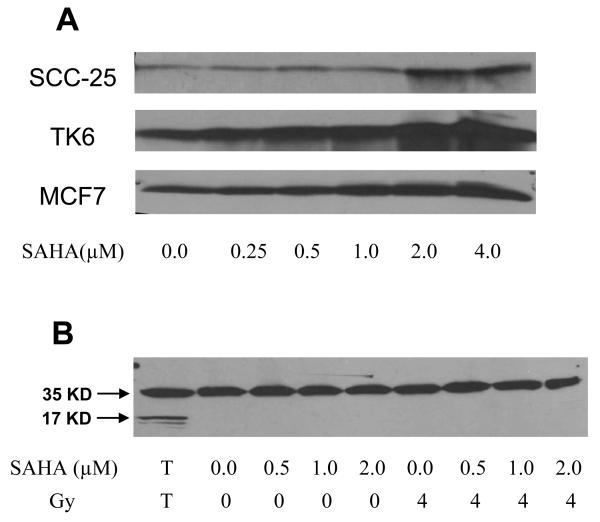
A gradual increase in the level of acetylation was observed with increase in SAHA concentration (Fig. 3A). For the MCF7 and TK6 cell lines, increased levels of acetylation of H4 histones were observed at low concentrations (0.5 μM and 1.0 μM) of SAHA. Equal loading of protein samples was confirmed by Coomassie blue staining of the protein gels (data not shown). From the Western blot data it can be concluded that pharmacologically relevant levels of SAHA can lead to acetylation of histones and hence a more ‘open’ chromatin structure.
Fig. 3.
A. Western blots showing that SAHA increases acetylation of H4 histones. SCC-25, MCF7 and TK6 cell lines were treated with 0.0 μM to 4.0 μM SAHA for 24 h and probed for acetylated H4 histones. B. Western blot showing SAHA does not induce apoptosis in SCC-25 cell line. SCC-25 cells were treated with 0.0 μM to 2.0 μM SAHA and then irradiated with 4 Gy. SAHA did not induce apoptosis either by itself or in combination with irradiation. T – TK6 cells treated with anti-CD95 antibody used as positive control.
SAHA modification of survival is independent of caspase3 activation
To assess the impact of apoptosis as a possible contributor to the radiosensitizing action of SAHA, levels of activated caspase 3 were measured. SCC-25 cells were irradiated with 4 Gy with or without pre-treatment with SAHA (0 μM to 2 μM). TK6 cells treated with anti-CD95 antibody were used as a positive control. For the positive control, both full length caspase-3 (35 kD) and the large fragment of caspase-3 (17 kD) resulting from cleavage activation were observed. For SCC-25 cells treated with SAHA and/or irradiation, no cleavage of caspase-3 was observed (Fig. 3B). This indicated that SAHA treatments up to 2 μM do not induce apoptosis in SCC-25 cells.
SAHA enhances 11q23 fragmentation
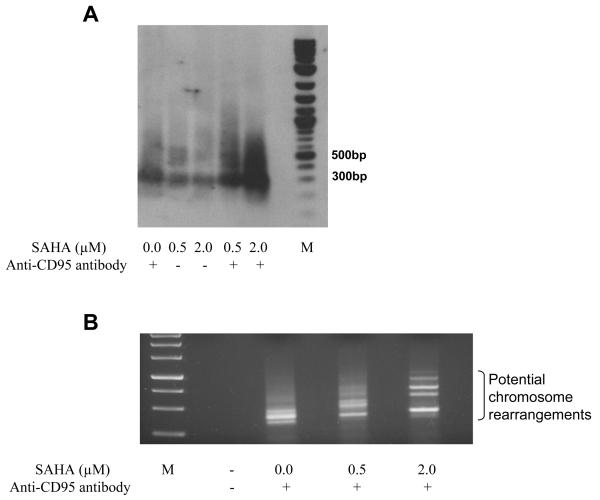
We hypothesized that the radiosensitizing activity of SAHA may be due to increased access of nucleases and/or free radicals to chromatin, secondary to SAHA modulation of histone acetylation, leaving chromatin in an open configuration. The interplay between SAHA treatment and endogenous nuclease cleavage was studied by examining breaks induced by anti-CD95 antibody within the11q23 region, using LM-PCR as the readout. It was observed that SAHA-only treatment did not alter the fragmentation pattern, but when used in combination with anti-CD95 antibody, there was a significant increase in the amount of breaks and their distribution within the target region (Fig. 4A). These data are consistent with greater accessibility of chromatin to endogenous nucleases subsequent to SAHA-induced chromatin relaxation.
Fig. 4.
Effect of SAHA on intensity of DNA breaks and potential aberrations. A. Southern blot showing the effect of SAHA on the intensity of breaks generated at 11q23 region by anti-CD95 antibody. SAHA did not increase the number of breaks but when used in combination with anti-CD95 antibody there was a significant increase in the amount and distribution of breaks. M – Molecular weight marker. B. IPCR results of TK6 cells treated with anti-CD95 antibody with or without pre-treatment with SAHA. The possible aberrations have been shown and each band was individually cloned and sequence analyzed. M – Molecular weight marker.
SAHA affects NHEJ dependent chromosome rearrangements
Radiation toxicity is linked to the creation of lethal lesions such as dicentric chromosomes; here we studied the role of SAHA in promoting such processes. After anti-CD95 antibody treatment potential chromosome rearrangements were captured at the 11q23 region using IPCR. Here, potential rearrangements are excised, circularized by T4 ligation and the entire construct PCR amplified using divergent primers to the known region (20). At the highest concentration of SAHA, the intensity of novel amplicons observed by gel electrophoresis is increased (Fig. 4B). For each treatment group, similar numbers of total PCR amplification products were captured by IPCR (Table 1). However, when compared to the total number of amplicons verified by sequencing, the percentage of rearrangements increased with SAHA concentration (Table 1). Those amplicons that were not verified as rearrangements contained 11q23 sequence only, with no partner DNA. The majority (14/18) of rearrangements showed microhomology at the site coe on indicating that the non-homologous end joining (NHEJ) repair pathway was likely involved (Table 2) (26). A reduction in base pair length of microhomology was observed at the highest SAHA concentration used. The average microhomology for the cells not treated with SAHA was 4.5±1.3, for 0.5 μM – 4.6±2.1, for 2.0 μM – 3.1±0.8. The rearrangements detected, recorded at the breakpoint junction here, may be extrapolated to illustrate a complete aberration, shown for one example in Fig. 5.
Table 1.
Summary of MLL aberrations
| SAHA Treatment |
Total aberrations |
DNA rearrangements |
Translocation (%) |
Microhomology (bp) |
|---|---|---|---|---|
| 0.0 μM | 14 | 4 | 28.57 | 4.5 ± 1.3 |
| 0.5 μM | 12 | 5 | 41.67 | 4.6 ±2.1 |
| 2.0 μM | 15 | 9 | 60.00 | 3.1 ± 0.8 |
All the three groups were treated with anti-CD95 antibody following pre-treatment with 0.0 μM (DMSO only), 0.5 μM and 2.0 μM SAHA respectively.
Table 2.
Details of Translocations detected following SAHA treatment
| SAHA Treatment |
Site | Chr. | MLL microhomology partner |
|---|---|---|---|
| 0.0 μM | 6766 | 6 | ATCTTCCCATGTTCAACTCCTCTT |
| 0.0 μM | 6807 | 20 | TGAGCCAAGGTCACACC |
| 0.0 μM | 6807 | 2 | CTGTTGTGAGCCAGGGCATT |
| 0.0 μM | 6894 | 5 | TGTTGTGAGCTCTCATATG |
| 0.5 μM | 6805 | 4 | TGTGAGTACAAACCAAG |
| 0.5 μM | 6852 | 1 | GCCCTCTGGAGGATGGTGGT |
| 0.5 μM | 6816 | 4 | TGTGAGTACAAACCAAGGAAAGTGG |
| 0.5 μM | 6807 | 14 | TGTTGTGAGCCTACGTTC |
| 0.5 μM | 6804 | 5 | TTGTGAATCTGGAA |
| 2.0 μM | 6719 | 5 | ATGGTTTTGCAATGG |
| 2.0 μM | 6805 | 1 | TTGTGAGATTCTAGC |
| 2.0 μM | 6806 | 1 | TCTGTTGTGAGCCACCAG |
| 2.0 μM | 6806 | 18 | TCTGTTGTGAGCCACTGCAC |
| 2.0 μM | 6807 | 13 | TGAGCCAAAACAGGA |
| 2.0 μM | 6789 | 2 | TGTGTATTGAGCCATG |
| 2.0 μM | 6759 | 20 | ACTTCTATCTATTTTATGTC |
| 2.0 μM | 6809 | 3 | GTGAGCCCTGTCTCCACT |
| 2.0 μM | 6807 | 10 | CTGTTGTGAGCAGTCGATG |
The site of break in the MLL-BCR for various rearrangements is listed. The microhomology between the MLL and partner chromosome is boldface. The sequence of the partner sequence is underlined.
Fig. 5.
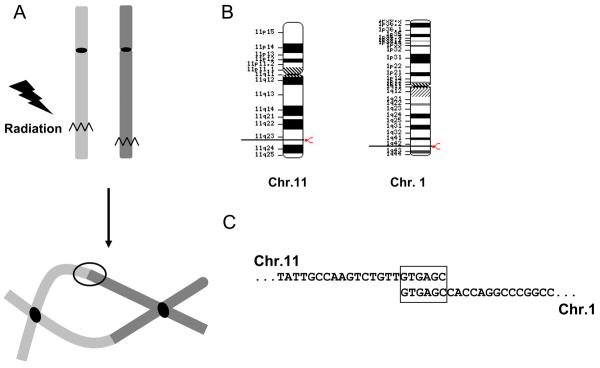
Diagrammatic representation of process involved in formation of DNA rearrangements. A) DNA-DSB in each of two chromosomes can lead to formation of dicentric chromosomes after a single round of cell division. B) DNA-DSB in Chromosome 11 and 1 and the subsequent DNA rearrangement captured by IPCR. C) Sequence of Chromosome 11 and 1 at the site of DNA rearrangement is shown as analyzed in these experiments and the microhomology between the two chromosomes is boxed.
Discussion
Although SAHA is being used in the clinic, its mechanism of action at a molecular level is still a matter for conjecture. Here we studied the effect of SAHA on both DNA damage induction and its subsequent conversion to unique DNA lesions, as well as its capacity to affect DNA repair relevant to cell survival.
In our study, we first looked at the acetylation capacity of SAHA in three cell lines. It was observed that SAHA can increase the acetylation of H4 histones at concentrations of 1 μM in all cell lines studied. This is clinically relevant as at the FDA approved dosage of SAHA of 400 mg once a day orally, the concentration of SAHA observed in patients is 1.2±0.53 μM (14). Thus pharmacologically relevant levels of SAHA were used to conduct subsequent in vitro studies. Following execution of clonogenic survival experiments in the presence or absence of SAHA, modest levels of radiosensitization were observed. Typically higher DEFs have been reported for SAHA but these studies have either used a much higher (2.5 μM) concentration of SAHA (5) or longer (3 days) incubation with SAHA (3). To address the mechanism of radiosensitization we looked at the ability to induce apoptosis, a known outcome of SAHA exposure in lymphocytes (27). To test this hypothesis, we probed for activated caspase 3 in SCC-25 by Western blot. A range of SAHA concentrations with or without irradiation did not induce any apoptosis in this epithelial derived cell line. Similar results were observed by Zhang et al when squamous carcinoma cell lines were treated with Trichostatin A, a HDACI that is structurally similar to SAHA, using flurometric assays for activated caspase 3 and 9; but this study did find increase in cellular necrosis (6). Hrzenjak et al have shown that SAHA induces caspase independent death of endometrial stromal sarcoma cells (28). This implies that SAHA can induce cell death by non-apoptotic pathways. Treatment with 0.5 μM SAHA did not have any effect on the cell cycle distribution of SCC-25 cells (data not shown), a finding also observed by others (28).
In order to understand the mechanism(s) driving radiosensitization by SAHA, SLDr experiments were conducted with the SCC-25 cell line. With this assay the ability of cells to successfully repair sublethal lesions are measured. The kinetics of increased survival measured by this technique has been linked to the kinetics of DNA double strand break (DNA-DSB) rejoining - the key feature in killing cells with ionizing radiation (29). When the time interval between the two radiation doses was more than 60 min, the SAHA treated cells showed an unusual collapse in effective repair, returning to levels at or below those observed without allowing time for DNA repair. In order to understand this observation, and its possible links to DNA breaks and their conversion to lethal lesions, we used molecular techniques to probe both the induction of a specific DNA break and its conversion into a unique rearrangement
To pursue this part of the study we made use of a known common fragmentation site at 11q23 that is frequently cleaved after exposure to genotoxic agents (20-22). This approach allowed us to analyze discrete molecular events rather than global effect of SAHA on DNA breaks introduced at random. It was observed, using LM-PCR to screen for the sites of specific DNA fragmentation, that both the distribution and intensity of breaks generated by anti-CD95 antibody treatment were significantly increased by pre-treatment with SAHA. We interpreted this result to indicate that a more ‘open’ configuration of chromatin promoted by SAHA increases DNA vulnerability to endogenous nucleases or free radical attack. To pursue the effects on DNA repair at the single cell level, a screen was carried out to measure the type of DNA rearrangements linked to radiation cell death.
The creation of chromosome rearrangements has historically been linked to the “mitotic catastrophe” route of cell death for irradiated cells (30). Such cells are characterized by the presence of chromosomal rearrangements, such as dicentrics, that physically limit partition at mitosis (31, 32). These observations have been included in the Theory of Dual Radiation action where individual DNA breaks recombine to form lethal lesions (33-36). The modern iteration of this theory is found in the analysis of cell survival curves and radiation response data using the linear quadratic equation – in particular the alpha and beta coefficients extracted from it (36). In order to model this process of aberration formation we studied the effect of SAHA on chromosomal aberrations at the same11q23 region. The advantage of studying aberrations at this fixed location is that it allows a study of the effect at the base pair level in a single cell, providing a better description of the process of aberration formation. Here we used IPCR following anti-CD95 antibody treatment to create a specific DNA break at 11q23 and found that SAHA treatment led to an increase in the efficiency of aberration detection. That is, more of the analyzed amplicons were verified by sequencing as actual chromosomal rearrangements, the remainder being derived from unaltered chromosome 11 DNA. Interestingly, increased amounts of SAHA also decreased the degree of microhomology at the chromosome breakpoints, microhomology is a signature feature of the error prone NHEJ repair pathway (Table 1) (26). The decrease in the number of microhomology base pairs with pre-treatment with SAHA may indicate that the open configuration facilitates the process of rearrangement formation, or at least modifies it from that observed in cells not exposed to SAHA. This would be at the stage where NHEJ repair enzymes, including Ku70/80, are loaded at the DNA break site (26). As an illustration of the impact of such rearrangements analyzed at the base pair level, a single dicentric rearrangement can be extrapolated from these molecular data and is presented schematically in Fig. 5. These data therefore indicate that SAHA has a role both at the level of DNA damage induction and at its subsequent repair or rearrangement. A decrease in the repair capacity by SAHA treatment has been suggested by the work of other groups where SAHA was able to alter the protein levels of genes involved in both homologous recombination and NHEJ repair (3, 5).
Our data indicates that SAHA can act as a radiosensitizer at pharmacologically relevant concentrations. Recent clinical trials of SAHA have indicated that it is well tolerated in humans (15-17). Although research has shown that SAHA may not be a good candidate as single drug treatment, it is a very promising candidate for combination therapy. Thus in the presence of agents such as irradiation and etoposide that introduce DNA-DSBs, SAHA may impact both DNA damage accumulation and repair, potentially leading to the creation of lethal lesions.
Acknowledgments
The Work was supported by NIH Grant CA105049 and from the Merck IISP program: 32974.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Notification
Conflicts of interest do not exist.
References
- 1.Tsai JY, Iannitti DA, Safran H. Combined modality therapy for pancreatic cancer. Semin Oncol. 2003;30:71–79. doi: 10.1016/s0093-7754(03)00273-2. [DOI] [PubMed] [Google Scholar]
- 2.Richon VM. Cancer biology: mechanism of antitumour action of vorinostat (suberoylanilide hydrosamic acid), a novel histone deacetylase inhibitor. British J Cancer. 2006;95:S2–S6. [Google Scholar]
- 3.Chinnaiyan P, Vallabhaneni G, Armstrong E, et al. Modulation of radiation response by histone deacetylase inhibition. Int J Radiat Oncol Biol Phys. 2005;62:223–229. doi: 10.1016/j.ijrobp.2004.12.088. [DOI] [PubMed] [Google Scholar]
- 4.Jung M, Velena A, Chen B, et al. Novel HDAC inhibitors with radiosensitizing properties. Radiat Res. 2005;163:488–493. doi: 10.1667/rr3345. [DOI] [PubMed] [Google Scholar]
- 5.Munshi A, Tanaka T, Hobbs ML, et al. Vorinostat, a histone deacetylase inhibitor, enhances the response of human tumor cells to ionizing radiation through prolongation of gamma-H2AX foci. Mol Cancer Ther. 2006;5:1967–1974. doi: 10.1158/1535-7163.MCT-06-0022. [DOI] [PubMed] [Google Scholar]
- 6.Zhang Y, Jung M, Dritschilo A. Enhancement of radiation sensitivity of human squamous carcinoma cells by histone deacetylase inhibitors. Radiat Res. 2004;161:667–674. doi: 10.1667/rr3192. [DOI] [PubMed] [Google Scholar]
- 7.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 8.Kim GD, Choi YH, Dimtchev A, et al. Sensing of ionizing radiation-induced DNA damage by ATM through interaction with histone deacetylase. J Biol Chem. 1999;274:31127–31130. doi: 10.1074/jbc.274.44.31127. [DOI] [PubMed] [Google Scholar]
- 9.Ju R, Muller MT. Histone deacetylase inhibitors activate p21(WAF1) expression via ATM. Cancer Res. 2003;63:2891–2897. [PubMed] [Google Scholar]
- 10.Bolden JE, Peart MJ, Johnstone RW. Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov. 2006;5:769–784. doi: 10.1038/nrd2133. [DOI] [PubMed] [Google Scholar]
- 11.Marks PA, Richon VM, Rifkind RA. Histone deacetylase inhibitors: inducers of differentiation or apoptosis of transformed cells. J Natl Cancer Inst. 2000;92:1210–1216. doi: 10.1093/jnci/92.15.1210. [DOI] [PubMed] [Google Scholar]
- 12.Munster PN, Troso-Sandoval T, Rosen N, et al. The histone deacetylase inhibitor suberoylanilide hydroxamic acid induces differentiation of human breast cancer cells. Cancer Res. 2001;61:8492–8497. [PubMed] [Google Scholar]
- 13.Marks P, Rifkind RA, Richon VM, et al. Histone deacetylases and cancer: causes and therapies. Nat Rev Cancer. 2001;1:194–202. doi: 10.1038/35106079. [DOI] [PubMed] [Google Scholar]
- 14.Mann BS, Johnson JR, Cohen MH, et al. FDA approval summary: vorinostat for treatment of advanced primary cutaneous T-cell lymphoma. Oncologist. 2007;12:1247–1252. doi: 10.1634/theoncologist.12-10-1247. [DOI] [PubMed] [Google Scholar]
- 15.Crump M, Coiffier B, Jacobsen ED, et al. Phase II trial of oral vorinostat (suberoylanilide hydroxamic acid) in relapsed diffuse large-B-cell lymphoma. Ann Oncol. 2008;19:964–969. doi: 10.1093/annonc/mdn031. [DOI] [PubMed] [Google Scholar]
- 16.Luu TH, Morgan RJ, Leong L, et al. A phase II trial of vorinostat (suberoylanilide hydroxamic acid) in metastatic breast cancer: a California Cancer Consortium study. Clin Cancer Res. 2008;14:7138–7142. doi: 10.1158/1078-0432.CCR-08-0122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Connor OA, Heaney ML, Schwartz L, et al. Clinical experience with intravenous and oral formulations of the novel histone deacetylase inhibitor suberoylanilide hydroxamic acid in patients with advanced hematologic malignancies. J Clin Oncol. 2006;24:166–173. doi: 10.1200/JCO.2005.01.9679. [DOI] [PubMed] [Google Scholar]
- 18.Shiozawa K, Nakanishi T, Tan M, et al. Preclinical studies of vorinostat (suberoylanilide hydroxamic acid) combined with cytosine arabinoside and etoposide for treatment of acute leukemias. Clin Cancer Res. 2009;15:1698–1707. doi: 10.1158/1078-0432.CCR-08-1587. [DOI] [PubMed] [Google Scholar]
- 19.Richon VM, Garcia-Vargas J, Hardwick JS. Development of vorinostat: current applications and future perspectives for cancer therapy. Cancer Lett. 2009;280:201–210. doi: 10.1016/j.canlet.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Betti CJ, Villalobos MJ, Diaz MO, et al. Apoptotic triggers initiate translocations within the MLL gene involving the nonhomologous end joining repair system. Cancer Res. 2001;61:4550–4555. [PubMed] [Google Scholar]
- 21.Betti CJ, Villalobos MJ, Jiang Q, et al. Cleavage of the MLL gene by activators of apoptosis is independent of topoisomerase II activity. Leukemia. 2005;19:2289–2295. doi: 10.1038/sj.leu.2403966. [DOI] [PubMed] [Google Scholar]
- 22.Libura J, Slater DJ, Felix CA, et al. Therapy-related acute myeloid leukemia-like MLL rearrangements are induced by etoposide in primary human CD34+ cells and remain stable after clonal expansion. Blood. 2005;105:2124–2131. doi: 10.1182/blood-2004-07-2683. [DOI] [PubMed] [Google Scholar]
- 23.Yang C, Betti C, Singh S, et al. Impaired NHEJ function in multiple myeloma. Mutat Res. 2009;660:66–73. doi: 10.1016/j.mrfmmm.2008.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Le H, Singh S, Shih SJ, et al. Rearrangements of the MLL gene are influenced by DNA secondary structure, potentially mediated by topoisomerase II binding. Genes Chromosomes Cancer. 2009 doi: 10.1002/gcc.20685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marschalek R. Etoposide-treatment and MLL rearrangements. Eur J Haematol. 2008;81:481–482. doi: 10.1111/j.1600-0609.2008.01139.x. author reply 483-485. [DOI] [PubMed] [Google Scholar]
- 26.Pastwa E, Blasiak J. Non-homologous DNA end joining. Acta Biochim Pol. 2003;50:891–908. [PubMed] [Google Scholar]
- 27.Frew AJ, Johnstone RW, Bolden JE. Enhancing the apoptotic and therapeutic effects of HDAC inhibitors. Cancer Lett. 2009;280:125–133. doi: 10.1016/j.canlet.2009.02.042. [DOI] [PubMed] [Google Scholar]
- 28.Hrzenjak A, Kremser ML, Strohmeier B, et al. SAHA induces caspase-independent, autophagic cell death of endometrial stromal sarcoma cells by influencing the mTOR pathway. J Pathol. 2008;216:495–504. doi: 10.1002/path.2434. [DOI] [PubMed] [Google Scholar]
- 29.Elkind MM, Sutton-Gilbert H, Moses WB, et al. Sub-lethal and lethal radiation damage. Nature. 1967;214:1088–1092. doi: 10.1038/2141088a0. [DOI] [PubMed] [Google Scholar]
- 30.Vakifahmetoglu H, Olsson M, Zhivotovsky B. Death through a tragedy: mitotic catastrophe. Cell Death Differ. 2008;15:1153–1162. doi: 10.1038/cdd.2008.47. [DOI] [PubMed] [Google Scholar]
- 31.Gisselsson D, Shao C, Tuck-Muller CM, et al. Interphase chromosomal abnormalities and mitotic missegregation of hypomethylated sequences in ICF syndrome cells. Chromosoma. 2005;114:118–126. doi: 10.1007/s00412-005-0343-7. [DOI] [PubMed] [Google Scholar]
- 32.Roninson IB, Broude EV, Chang BD. If not apoptosis, then what? Treatment-induced senescence and mitotic catastrophe in tumor cells. Drug Resist Updat. 2001;4:303–313. doi: 10.1054/drup.2001.0213. [DOI] [PubMed] [Google Scholar]
- 33.Darroudi F, Bezrookove V, Fomina J, et al. Insights into the sites of X ray and neutron induced chromosomal aberrations in human lymphocytes using COBRA-MFISH. Radiat Prot Dosimetry. 2002;99:189–192. doi: 10.1093/oxfordjournals.rpd.a006758. [DOI] [PubMed] [Google Scholar]
- 34.Schmid E, Braselmann H, Nahrstedt U. Comparison of gamma-ray induced dicentric yields in human lymphocytes measured by conventional analysis and FISH. Mutat Res. 1995;348:125–130. doi: 10.1016/0165-7992(95)00056-9. [DOI] [PubMed] [Google Scholar]
- 35.Schroder H, Heimers A. Chromosome aberrations induced in human lymphocytes by in vitro and in vivo X-rays. Mutat Res. 2002;517:167–172. doi: 10.1016/s1383-5718(02)00067-0. [DOI] [PubMed] [Google Scholar]
- 36.Zaider M, Rossi HH. Dual radiation action and the initial slope of survival curves. Radiat Res Suppl. 1985;8:S68–76. [PubMed] [Google Scholar]