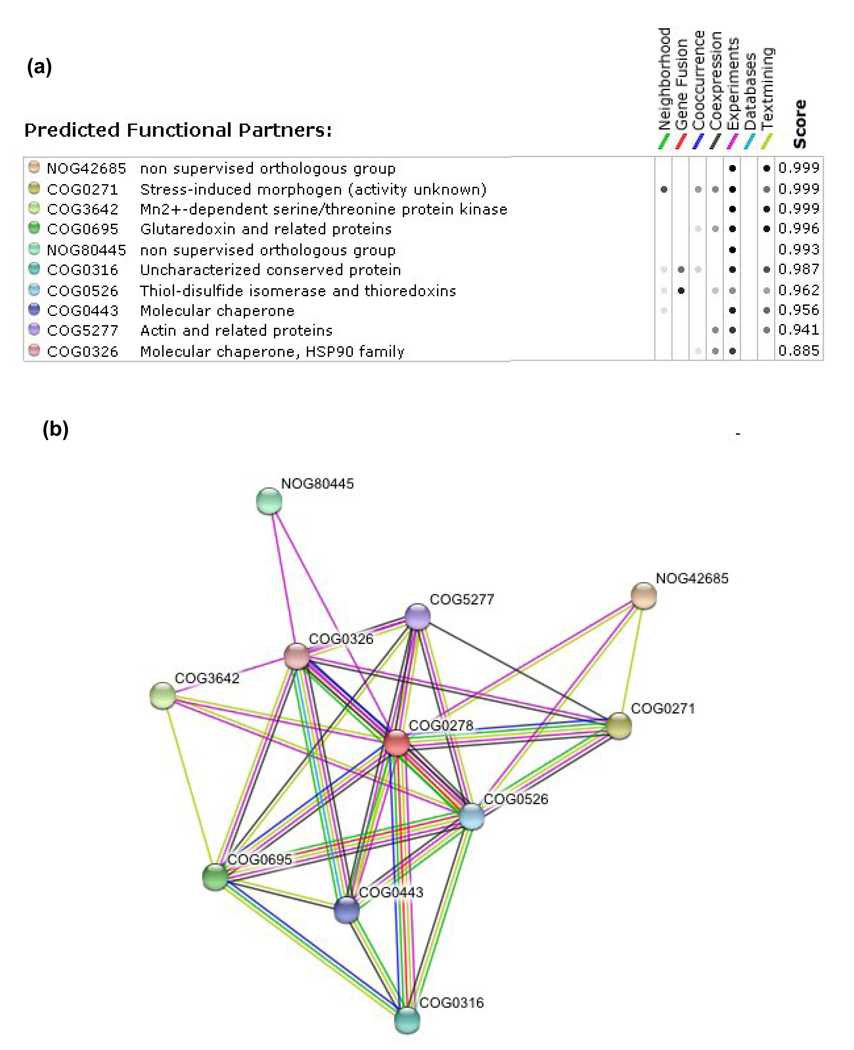
Figure 4. Putative CGFS Grx target proteins retrieved using the STRING webtool.
An in silico genomic analysis based notably on gene fusion, gene clustering, gene co-occurrence and gene co-expression, and combined to experimental data, was used to predict putative functional or physical interacting proteins. The 10 most confident putative CGFS Grx-interacting proteins (COG0278) (a) and their representation as a Grx network (b) are listed along with the 7 criteria which were used in the analysis. In this network, the number of connections, which represents the different criteria used, is used as a correlation for the strength of the interaction, unless experimental data exist for such interaction. NOG42685 corresponds to Aft1p transcription factor, COG0271 to BolA proteins, COG0316 to IscA/HesB/YfhF scaffold proteins, COG0443 to HscA/Hsp70/Ssq1p chaperones, COG0326 to HscB/Hsp90/Jac1p chaperones, COG3642 to a family of serine/threonine protein kinases with Bud32p as representative, COG0695 to class I Grxs, NOG80445 to phosphatidylinositol 4,5-bisphosphate protein, COG0526 to Trxs with Grx3/4p and plant GrxS17 as representatives and COG5277 to actin.