Figure 1.
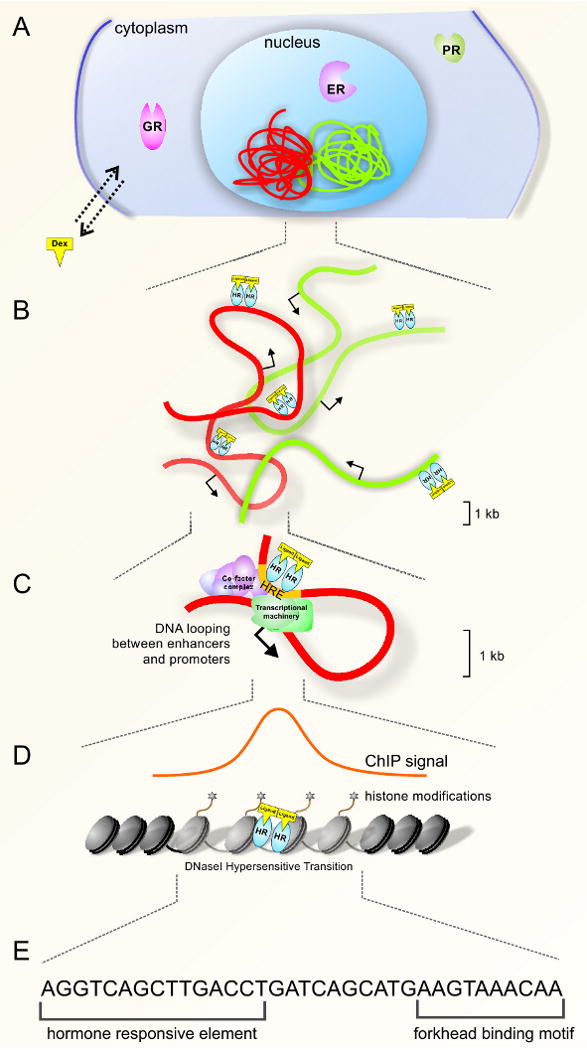
Epigenomic mechanism of nuclear receptor action. (a) Chromatin is organized into higher-order structures as chromosomes (red and green structures) which are non-randomly positioned in the nucleus. Nuclear receptors reside in the cytoplasm or nucleus in the unliganded state. Upon lipophilic ligand binding to receptors, the receptor undergoes a conformational change to mediate DNA binding and transcriptional regulation. (b) Hormone receptors (HR), such as estrogen receptor (ER), glucocorticoid receptor (GR) and progesterone receptor (PR), bind sequences on DNA, predominantly at distal regulatory elements. Arrows indicate direction of transcription. (c) Nuclear receptor binding to distal regulatory elements may involve long-range interactions including looping to facilitate recruitment of enhancers to promoter regions. (d) Analysis of receptor binding by chromatin immunoprecipitation (ChIP) coupled with tiled microarrays (ChIP-chip) or sequencing (ChIP-Seq) permits genome-wide resolution of receptor binding. Binding sites are associated with modifications of histone tails and chromatin accessibility, monitored by transitions in sensitivity to nucleases such as DNaseI. (e) Motif analysis of nuclear receptor binding sites identifies nuclear receptor response elements, but also motifs for transcription factors that may play important roles in nuclear receptor function.
