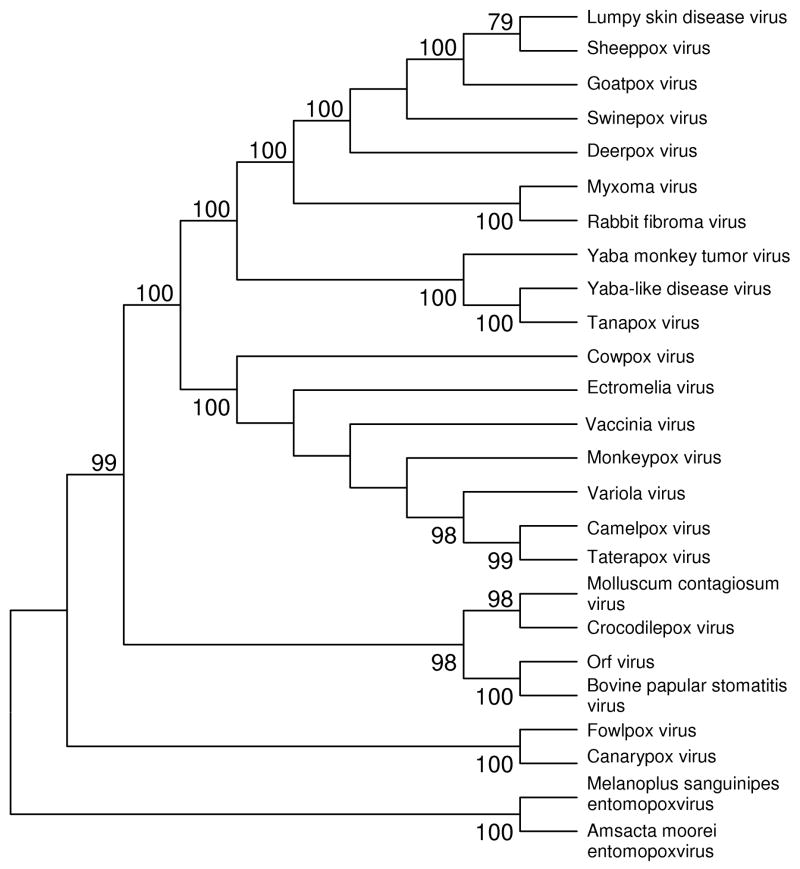
Figure 1.
NJ tree (topology only) of poxviruses based on concatenated amino acid sequences from 29 conserved orthologous proteins (13, 475 aligned sites). The tree was constructed on the basis of the JTT amino acid distance, assuming that rate variation among sites follows a gamma distribution (shape parameter a = 0.86). Numbers on the branches represent percentages of 1000 bootstrap samples supporting each branch; only values ≥ 50 % are shown. For accession numbers of genome sequences, see Supplementary Table S1.