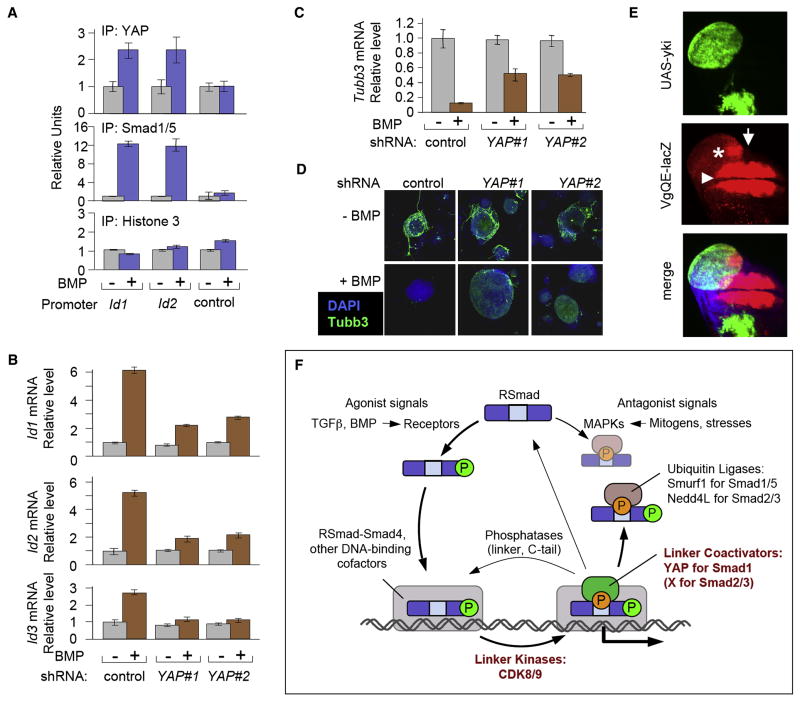
Figure 7. YAP enhances BMP-Smad responses in different biological contexts.
(A) Chromatin immunoprecipitation of BMP-stimulated mESCs using antibodies against YAP and Smad1/5. BMP-responsive regions of Id1 and Id2 and an unresponsive control region of Id1 (control) were analyzed by qRT-PCR as in Figure 2D. (B) Wild-type YAP-knockdown mESCs were stimulated with BMP4 for 1 h. Total RNA was analyzed by qRT-PCR. Data show the mean ± S.D of quadruplicates and are representative of two independent experiments. (C) Gene expression analysis of the neural differentiation marker β-III tubulin (Tubb3), in differentiating wild-type and YAP-knockdown mESCs. Cells were cultured in N2B27 supplemented media in the presence or absence of BMP, harvested five days later and total RNA was subjected to qRT-PCR analysis for Tubb3 expression as in (B). (D) Confocal images of mESCs treated as in (C) and subjected to immunofluorescence staining with an anti-Tubb3 antibody (green) and DAPI counterstaining (blue). Tubb3 staining detects neuronal differentiation. (E) vgQE-lacZ expression (red) in Drosophila wing imaginal disc containing Yorkie-overexpressing clones (marked by GFP+). Confocal z-stack through the wing imaginal disc was projected into a single plane. Note that the ectopic vgQE-lacZ expression (asterisk) is discontinuous with the endogenous vgQE-lacZ expression domain (arrowhead) and only observed close to the anterior-posterior compartment boundary (arrow). (F) The Smad signaling cycle. Receptor-bound BMP and TGFβ ligands induce C-tail phosphorylation of R-Smads, which then accumulate in the nucleus. Nuclear R-Smads complexed with Smad4, bind to the regulatory elements of target genes and interact with other DNA binding cofactors (shown as a grey box), becoming linker-phosphorylated by CDK8/9 at some point during this process. This facilitates Smad1 binding to YAP, which is required for efficient transcription of BMP target genes. Linker phosphorylated Smad2/3 is thought to activate transcription of TGFβ target genes in a similar manner, but through as yet unknown cofactors. After fulfilling their transcriptional role, Smads may be targeted by linker and tail phosphatases that reset them to their ground state or linker phosphatases only that may allow further Smad activity. Alternatively, Smads can be recognized in a phospho-linker dependent manner, by ubiquitin ligases, such as Smurf1 in the case of Smad1, and Nedd4L in the case of Smad2/3, resulting in their eventual degradation. The R-Smad linker regions can also be phosphorylated by MAPK pathway kinases in response to antagonists, such as FGF, EGF, or stress signals, which ultimately leads to Smad degradation. By providing a platform for the regulation of the different and often opposing fates of the Smads, linker phosphorylation represents a focal point in the Smad signaling cycle.