Fig. 3. cJun co-localized to the G-CRE with CREB1 and ATF-2.
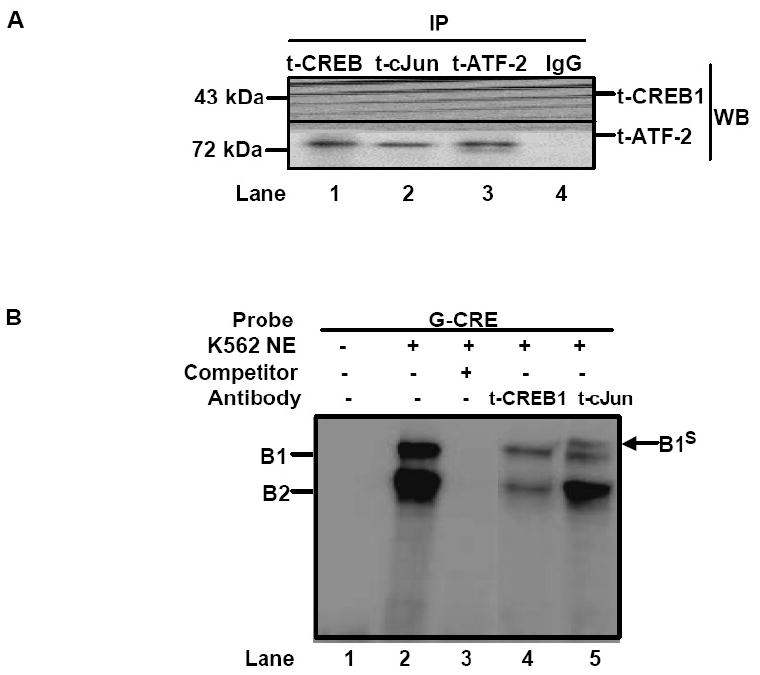
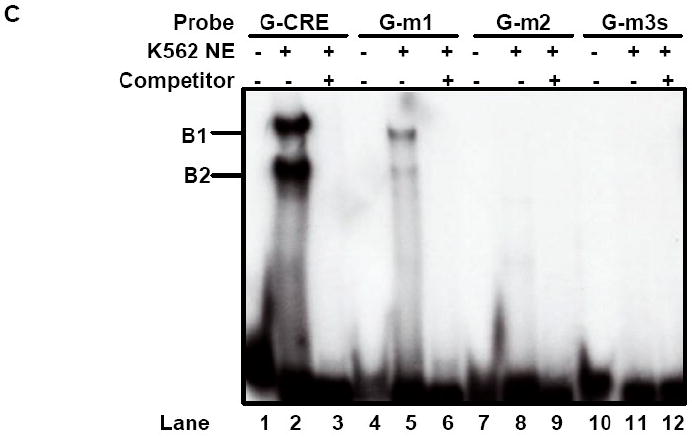
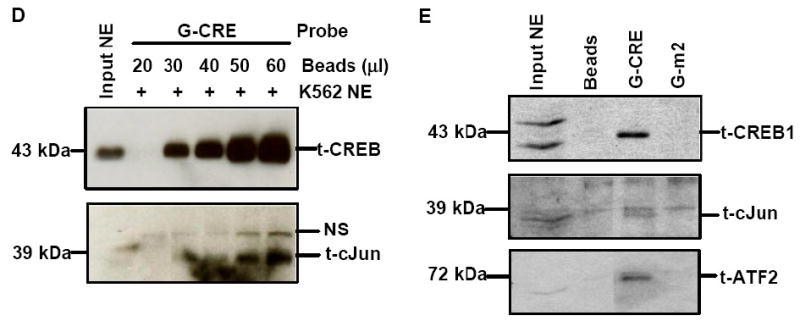
A) Immunoprecipitation (IP) reactions were performed with 200-300 μg of K562 nuclear extracts and t-CREB1, t-cJun, t-ATF-2 and IgG antibodies and then western blot (WB) was performed with separate membranes and the antibody shown to detect protein-protein interaction (See Materials). Note the presence of a protein band when the IP and WB antibody are identical serving as a positive control. IP with IgG served as a negative control and did not produce a protein band as expected. B) Electrophoretic mobility shift assay (EMSA) was performed with K562 cell nuclear extracts (NE) and the G-CRE probe. Different reactions in the absence (Lane 1) or presence of 8μg of K562 NE (Lanes 2-5) are shown. Supershift assays were performed with t-CREB1 and t-cJun antibodies. C) EMSA was performed under the same conditions as described in Panel B except reactions with the mutant probes G-m1 (G/A), G-m2 (AC/TG) and G-m3 with a scrambled G-CRE (tctctgta) were included. Competition reactions were completed in the presence of NE and cold oligonucleotide (++) to determine specificity of binding. D) Promoter pull-down reactions were performed with 2 μg of 5′biotinylated G-CRE probe and 300 μg of nuclear extract prepared from K562 cells. Increasing amounts of streptavidin beads (Beads) were analyzed to test efficiency of complex pull-down. The proteins bound to the G-CRE were detected by western blotting with t-CREB1 and t-cJun antibody (see Materials). Nuclear extract (Input NE) was used as a positive control. A non-specific (NS) protein band was observed on the t-cJun western blot. E) Promoter pull-down was performed with 5′biotinylated G-CRE and G-m2 (AC/TG) mutant probes as described in Panel D except 20 μl of streptavidin agarose beads were used in all reactions. Western blot analysis was also performed with total ATF-2 (t-ATF-2) antibody. A negative control reaction run in the absence of biotinylated probes (Beads) is shown.
