Figure 1. Identification of PDLIM5 as a SPAR interacting protein.
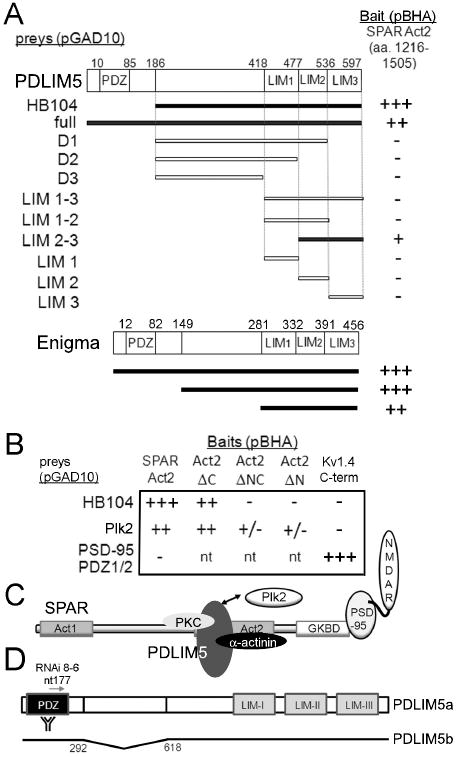
(A) Yeast 2-hybrid screen (Y2H) was performed using as bait SPAR Act2 domain in vector pBHA. Interaction strength for all Y2H assays was scored in terms of β-gal activity (+++, 0-15 min; ++, 15-30 min; +, 30 min to 2 hrs; +/-, 2 hrs to overnight; -, not detected; nt, not tested). Region of PDLIM5 encoded by positive clone HB104 and deletion analysis of PDLIM5 are shown below schematic diagram of PDLIM5 primary structure. Bottom, regions of Enigma tested in Y2H assay are shown below schematic diagram of Enigma primary structure. Numbers shown are amino acid residues; interacting clones are highlighted in black. (B) Deletion analysis of SPAR Act2. Domain constructs lacking the N terminal portion of Act2 (ΔC), the C terminal portion (ΔN), or both (ΔNC) were assayed by Y2H for interaction with PDLIM5 clone HB104. Plk2 in vector pGAD10 is used as a positive control for interaction with Act2, and PSD-95 PDZ1/2 domain in pGAD10 is used as negative control. Kv1.4 C-terminus is used as a positive control for PSD-95 PDZ1/2 interaction. (C) Schematic of SPAR interactions. SPAR domains shown are Act1 and 2, actin-associated domains 1 and 2; GKBD, guanylate kinase binding domain that interacts with the guanylate kinase-like domain of PSD-95. NMDAR, NMDA receptors. Double arrow denotes PDLIM5 and Plk2 interaction with the same region of SPAR. (D) Schematic of PDLIM5 reagents used in the study. PDLIM5a (full-length) and PDLIM5b (deletion of nt 292-618), position of RNAi 8-6, and PDZ region used as immunogen for PDLIM5 rabbit antibodies are shown.
