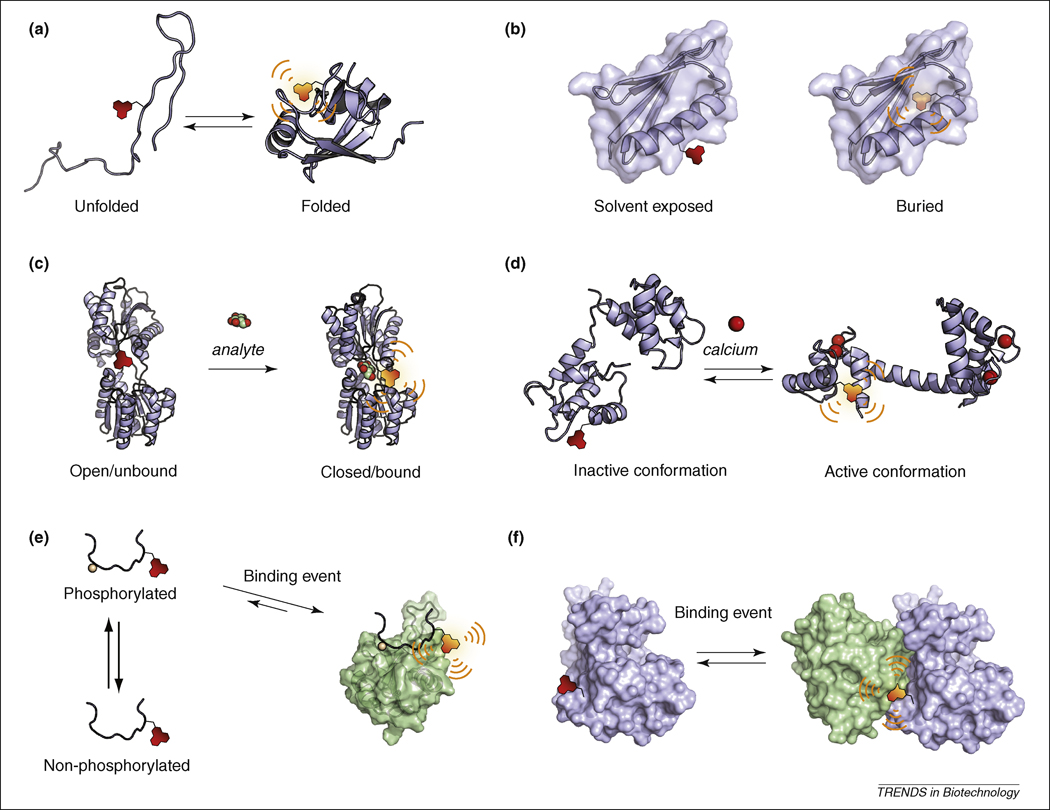
Figure 3. Applications of solvatochromism for biomolecular protein studies.
(a) Folding studies. (b) Mapping of a protein local environment and/or solvatation. (c) Sensors for small molecule analytes. The sensors are generally designed by exploiting protein domains that intrinsically bind a select analyte resulting in fluorescence change due to a conformational change in the protein or the displacement of the fluorophore (illustrated here with the d-glucose/d-galactose-binding protein from E. Coli, 2FW0 for the apo state and 2FVY for the glucose-bound state). (d) Reporting protein structural changes. In response to a signaling event, the protein of interest will undergo a conformational change that corresponds to a different functional state, which may be monitored by an appropriately positioned solvatochromic fluorophore (illustrated with calmodulin, 1DMO apo state and 1UP5 calcium-bound state). (e) Fragment- or peptide-based probes for the monitoring of protein interactions. A minimal fragment of one of the binding partners can be labeled with a solvatochromic fluorophore to report interactions. In the case of transient interactions, the signaling event that will induce the interaction can affect either one of the partners thus providing information on function or activity. This approach is illustrated here with the Crk SH2 domain (1JU5) that binds to phosphopeptides sequences and that can be used to report either the activity of a kinase or the phosphorylated state of a substrate. (f) Reporting protein-protein interactions. A similar approach to panel (e) can be applied to full-length proteins instead of fragments. In this case advantage can be taken of the larger interaction interface between two proteins compared to peptide-based probes that do not adopt secondary and tertiary structures.