Abstract
Throughout the animal kingdom striated muscle contraction is regulated by the thin filament troponin-tropomyosin complex. Homologous regulatory components are shared among vertebrate and arthropod muscles; however unique protein extensions and/ or components characterize the latter. The Troponin T (TnT) isoforms of Drosophila indirect flight and tarantula femur muscle for example contain distinct C-terminal extensions and are ~20% larger overall than their vertebrate counterpart. Using electron microscopy and three-dimensional helical reconstruction of native Drosophila, tarantula and frog muscle thin filaments we have identified species-specific differences in tropomyosin regulatory strand densities. The strands on the arthropod thin filaments were significantly larger in diameter than those from vertebrates, although not significantly different from each other. These findings reflect differences in the regulatory troponin-tropomyosin complex, which are likely due to the larger TnT molecules aligning and extending along much of the tropomyosin strands’ length. Such an arrangement potentially alters the physical properties of the regulatory strands and may help establish contractile characteristics unique to certain arthropod muscles.
Keywords: Actin, troponin, tropomyosin, electron microscopy, indirect flight muscle, contraction
Introduction
ATP-dependent cyclical interactions between myosin-containing thick filaments and actin-containing thin filaments drive muscle contraction in all animals. In resting striated muscle of vertebrates and higher invertebrates, the thin filament troponin-tropomyosin regulatory complex inhibits contraction by blocking high affinity myosin binding sites on actin [1–3]. Upon activation, tropomyosin moves azimuthally over the filament away from these sites in a stepwise manner as a result of Ca2+ binding to troponin followed by myosin crossbridge binding to actin [4–6]. Biochemical and structural data suggest that initial crossbridge binding evokes allosteric effects on thin filaments involving a propagated opening of myosin binding sites along the filaments leading to cooperative activation of contraction [1,3–5]. Thus, both elevated Ca2+ levels and crossbridge binding are required for complete activation of muscle contraction.
Tropomyosin is an elongated 42 nm coiled-coil protein that assembles end-to-end to form continuous strands which run along the entire length of the F-actin helices [7–9]. Each tropomyosin molecule possesses seven successive quasi-repeating motifs and is designed to bind to and span seven neighboring actin monomers along the filaments [10,11]. In vertebrate striated muscles, the C-terminal two-thirds of tropomyosin associates with specific domains of troponin, an asymmetric protein complex comprised of TnT (37 kDa), TnI (24 kDa), and TnC subunits (18 kDa) [9,12–15]. Thus, a single thin filament “regulatory unit” consists of seven actin subunits controlled by a single troponin-tropomyosin complex. TnC serves as the Ca2+ sensor of the troponin complex. Ca2+ binding to TnC relieves the inhibition of actomyosin interactions imposed by TnI. Elongated TnT (19 nm in vertebrates) binds the entire complex to tropomyosin [9,12,13,16]. The N-terminal “tail” of vertebrate TnT, predicted to be alpha helical, extends along the C-terminal half of tropomyosin as well as the end-to-end contacts between neighboring tropomyosin molecules [1,9]. The C-terminal portion of TnT forms part of the structural scaffold that supports TnC and TnI in the globular core domain of troponin [17,18]. TnT may also contribute to regulating actomyosin ATPase activity and to establishing the cooperative activation of the thin filament [1] by increasing the inhibition of actomyosin ATPase activity in the absence of Ca2+ and increasing activation in the presence of Ca2+ [19,20].
The components of thin filaments in invertebrates and vertebrates are highly homologous. For example, well-studied Drosophila melanogaster indirect flight muscle (IFM) and tarantula leg muscle thin filaments contain actin, tropomyosin, TnC, TnT and TnI homologues [21,22]. However, arthropod muscles often express troponin subunits with discrete extensions. For example, the TnT homologues in Drosophila IFM (46 kDa) and in tarantula muscle (major isoform 43 kDa) share 51% identity with each other and are ~20% larger than their vertebrate counterparts [22–24]. In IFM, the TnT subunit contains a ~136 amino acid long C-terminal extension that is highly acidic [23,24] and may account for the observed Ca2+ binding properties of this species of TnT [24]. The major tarantula TnT isoform also possesses an acidic C-terminal extension, though shorter than that expressed in Drosophila IFM [22]. Furthermore, Drosophila IFM expresses a TnI isoform (30 kDa) larger than that found in vertebrate or in tarantula (major isoform 24 kDa) skeletal muscle [22,25,26]. The ~6 kDa difference is predominately attributed to expression of an IFM specific exon that encodes an N-terminal proline- and alanine-rich extension. Additionally, two different “heavy” tropomyosin molecules (TmH), which contain a ~250 amino acid proline-rich carboxy-terminal domain beyond the normal tropomyosin sequence, are expressed in the IFM (but not in insect synchronous muscles or in other arthropod muscles) [27]. Stoichiometric measurements and protein-protein interaction studies of the TmH isoforms with standard IFM tropomyosin suggested that the TmH N-termini are integrated into the thin filament structural unit as tropomyosin homo- or heterodimers [27]. Immuno-electron microscopy indicated that TmH is homogeneously distributed along with normal tropomyosin on IFM thin filaments [28,29]. The proline-rich, C-terminal portion of TmH was modeled as an extended globular “spring” projecting out from the thin filament in a region close to the globular domain of the troponin complex [29].
EM and three-dimensional (3D) helical reconstruction are powerful imaging techniques for high resolution analysis of native and reconstituted thin filaments. They have played a key role in revealing the reorganization of the troponin-tropomyosin regulatory components that occurs in response to Ca2+ [30–32]. However, helical reconstruction treats all densities associated with F-actin as if they are identical on each monomer along the filament, and the average density contribution of discretely distributed structures, such as troponin complexes, can potentially merge into continuous structures (like tropomyosin), and not be easily delineated [33]. In particular, extended proteins such as TnT that run parallel to tropomyosin along much of their length, may not be discretely detected or resolved separately from the tropomyosin regulatory strands [33]. Therefore, tissue and phylogenetic differences in tropomyosin and/ or TnT molecules could make different contributions to the apparently well-defined actin monomers or the tropomyosin strands. Thus, in addition to comparing differences in thin filament structure under defined chemical conditions that correspond to different physiological states from the same organism, EM and helical reconstruction can be used to compare thin filaments under a single defined state from different organisms. This allows direct comparison and statistical evaluation of potential differences that exist in thin filaments possibly due to unique or distinct arrangements of regulatory components.
Previous imaging of Drosophila IFM thin filaments suggested the occurrence of larger tropomyosin regulatory strands when compared to those present in other organisms [31]. However, these inferences were not quantified, nor were the sources of these differences identified. In the present study, we have compared the structures of Drosophila, tarantula and frog thin filaments and find that the extra density associated with arthropod tropomyosin strands is due to the larger TnT isoform. We conclude that the arthropods’ negatively charged C-terminal TnT extensions align and extend along tropomyosin and do not project outward or away from the thin filament. Such an orientation could alter the regulatory strands’ physical properties and likely contributes to the muscles’ contractile characteristics.
Materials and Methods
Thin filament isolation
IFM thin filaments were isolated from the thoraces of Mhc7 Drosophila according to [31]. Tarantula thin filaments were purified from femur muscles as in [34]. Vertebrate thin filaments were extracted from Sartorius muscles of Rana pipiens according to [35] with minor modifications. Frogs were decapitated and the sartorius muscles were isolated. During the dissection the tissue was continually rinsed with cold rigor buffer (100 mM NaCl, 3 mM MgCl2•6H2O, 0.2 mM EGTA, 5 mM PIPES, 5 mM NaH2PO4•H2O, 1mM NaN3, 0.5 mM PMSF, pH 7.0). Both muscles from individual frogs were placed in 45 ml of skinning solution and slowly agitated overnight on ice. Superficial muscle layers were dissected, rinsed in 10 ml of rigor buffer and homogenized in 5 ml of rigor buffer with an omnimixer for 15 seconds and cooled for 15 seconds on ice. The blending and cool down steps were repeated three times. The homogenate was filtered through cheesecloth and was spun at 13,000 g for 15 min to pellet myofibrils. The myofibrils were washed and centrifuged at 13,000 g for 15 min three times. The final “washed” myofibrils were resuspended in 200 μl of relaxing solution (rigor buffer supplemented with 5mM EGTA) and gently homogenised to dissociate thick and thin filaments. The sample was spun at 15,000 g for 20 min to pellet thick filaments and the supernatant containing thin filaments was diluted 10 – 20 fold with relaxing solution immediately prior to grid preparation. All thin filaments preparations were maintained in EGTA to ensure low Ca2+ conditions.
Electron microscopy and 3D reconstruction of thin filaments
In this work, only low Ca2+ thin filament images were analyzed. This is because the variance associated with the relative positions of F-actin and tropomyosin densities in maps of 3D reconstructions of low Ca2+ data is less than those in high Ca2+.
5 μl of Ca2+-free filament suspension was applied to thin carbon films that were layered over holey carbon films supported on 400 mesh copper electron microscope grids. The thin filaments were negatively stained with 1% (w/v) uranyl acetate and the grids were dried as previously [5,36,37]. Images of filaments lying on thin carbon over the holes were recorded under low dose conditions at a magnification of 60,000X with a Philips CM 120 electron microscope operated at 80 kV. Images were digitized on a Zeiss SCAI scanner. Helical reconstruction of negatively stained thin filaments, which resolves actin monomer structure and tropomyosin strands, was carried out according to standard methods [5,36,38]. Helical projections were calculated from the 3D reconstructions of the thin filaments by projecting the component densities in the reconstructions down the long-pitch actin helical paths onto a plane perpendicular to the thin filament axis [30,38]. The resulting maps thus show axially averaged positions of actin and tropomyosin densities, appearing symmetrically on both sides of the filament. These projections are well suited for aligning and comparing the averaged densities from different species. A Student’s t test was performed for every component density of the averaged 3D maps [36,39]. Only regions whose average density was greater than baseline density in each map at better than the 99.95% confidence level are shown. Difference density analysis [30] between aligned maps from different organisms can reveal phylogenetic differences among their tropomyosin strand densities. The difference maps were evaluated by a Student’s t test [40].
Results/ Discussion
Electron microscopy and 3D reconstruction of low Ca2+-treated thin filaments
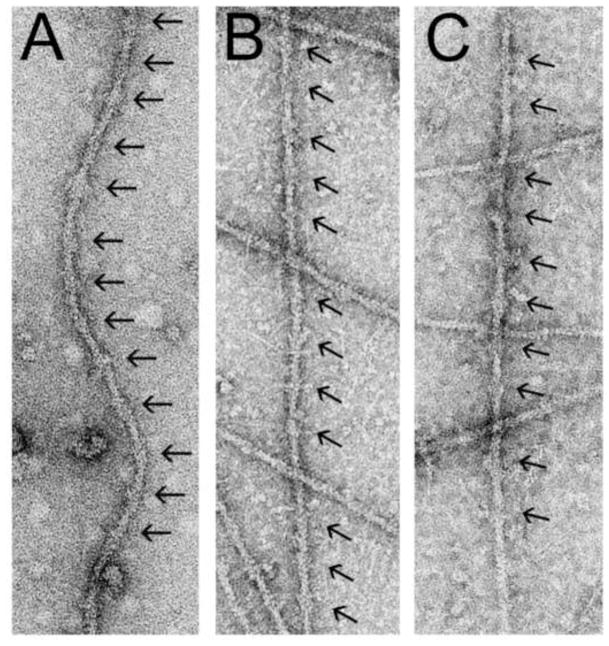
Thin filaments were isolated from Drosophila indirect flight, tarantula leg and frog Sartorius muscles and maintained in low Ca2+ conditions (Figure 1). Electron micrographs of negatively stained thin filaments showed characteristic double-helical arrays of actin subunits together with well-resolved, periodic troponin complexes (Figure 1).
Figure 1.
Electron micrographs of negatively stained native thin filaments from Drosophila melanogaster IFM (A), tarantula femur (B) and Rana pipiens sartorius muscle (C). Note the regions of thin filaments possessing large troponin complexes located every ~38.5 nm (black arrows).
Density maps were calculated from the averaged Fourier transform layer-line data obtained from 15 IFM thin filaments, 15 Rana sartorious muscle thin filaments and 10 tarantula leg muscle thin filaments (Table 1). Overall this translates into maps computed from data obtained from at least 60 thin filament regulatory units per species.
Table 1.
Summary of parameters used in image analysis
| Sample | n | ψ(°) | Δ ψ(°) | Significancea |
|---|---|---|---|---|
| EGTA-treated Drosophila IFM Thin Filamentsb | 15 | 58.0 +/− 5.6 | 23.9 +/− 6.6 | >99.95% |
| EGTA-treated Tarantula Thin Filaments | 10 | 53.2 +/− 7.1 | 27.1 +/− 8.0 | >99.95% |
| EGTA-treated Rana Thin Filaments | 15 | 58.1 +/− 7.9 | 20.7 +/− 7.7 | >99.95% |
n, number of filaments averaged to generate the layer-line data sets and reconstructions.
ψ(°), average phase residual (degrees) +/− standard deviation between individual filament layer-line data sets and the average of the data, a measure of the accuracy of the filament alignment.
Δ ψ(°), average up-down phase residuals (degrees) +/− standard deviations of individual layer line data sets, a measure of the filament polarity.
the statistical significance of actin and tropomyosin densities, determined by Student’s t-test.
from [31]
Helical projections, which reveal the axially averaged positions of tropomyosin strands relative to actin, were calculated from the 3D reconstructions of the thin filaments by projecting the component densities in the reconstructions down the long-pitch actin helical paths onto a plane perpendicular to the thin filament axis (Figure 2). The resulting maps thus show actin and the tropomyosin regulatory strands, which appeared symmetrical on both sides of the filament. The projections revealed typical two-domain actin monomers and tropomyosin strands which clearly made contact with the inner edge of the outer domain of actin. All actin and tropomyosin regions of each individual map possessed density that was significantly different from baseline density at greater than the 99.95% confidence level (Table 1).
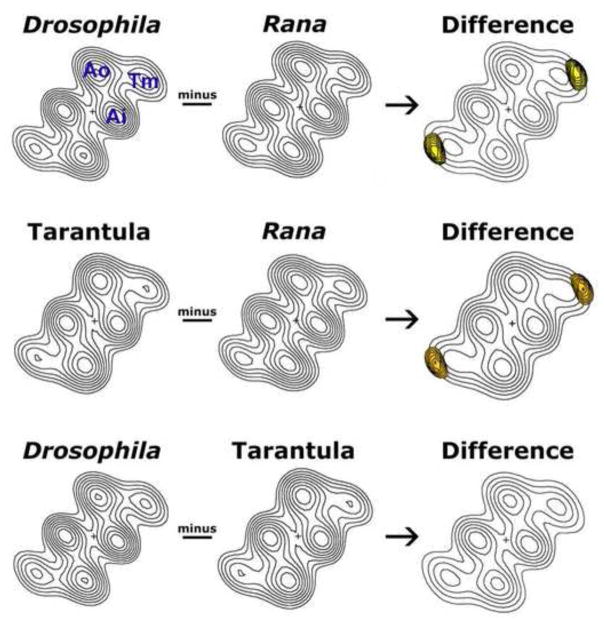
Figure 2.
Difference density analysis of 3D reconstructions of native thin filaments from Drosophila melanogaster IFM, tarantula femur muscle and Rana pipiens sartorius muscle. Helical projections were calculated from the 3D reconstructions of the thin filaments. The maps show axially averaged positions and dimensions of actin and tropomyosin densities. Ao = outer domain of actin, Ai = inner domain of actin, Tm = tropomyosin. In the absence of Ca2+, tropomyosin strands clearly contact the outer domain of actin. These projections are well suited for aligning and comparing the average densities along thin filament lengths, from different species. Subtraction of the densities of the Rana map from either Drosophila or tarantula maps reveals the presence of difference peaks (yellow) within the tropomyosin strands. These peaks are shown superimposed on the Drosophila or tarantula helical projections (displayed with limited contours for clarity). The differences are significant at greater than the 95% confidence level. No significant differences were detected between the Drosophila and tarantula tropomyosin strands.
Unique species-specific densities are statistically significant
Difference maps were computed by point-by-point subtraction of densities in one map from another (Figure 2). This showed that well-defined and invariant F-actin densities were common to all maps, illustrating the high degree of inter-species F-actin structural homology. However, the diameter of the densities associated with tropomyosin strands in the Drosophila and tarantula reconstructions were distinctly larger than those of the frog one. Statistical tests demonstrated that these differences (Figure 2, yellow peaks) were significant at greater than the 95% confidence level. However, comparison of the Drosophila and tarantula strands revealed no significant differences between these two sets of data despite the absence of TmH and the N-terminal TnI extension in the latter [22,27]. Thus, it is unlikely that the extra density found along the arthropod thin filaments is due either to the TmH proline-rich C-terminal region, or due to TnI proline/ alanine-rich N-terminal sequences, specifically aligning linearly along tropomyosin. This is consistent with the model of Reedy et al. [29], which suggested the proline-rich, C-terminal extended “spring” of TmH projects outward and away from the IFM thin filament. Similarly, we propose that the N-terminal TnI moiety also projects away from the filament axis, with a potentially random distribution that precludes the extension from adding extra density to IFM tropomyosin. Since the TnC isoforms among all 3 species analyzed here are similar in size [22,41,42], our results indicate that it is solely the elongated TnTs (and most likely the acidic C-terminal extensions) found in arthropods that makes a greater contribution to strand density than does vertebrate TnT. However, subtle contributions made by TmH and/or TnI on IFM filaments may not have been resolved adequately by our method.
Differences in Drosophila tropomyosin regulatory strand density correlate with specialized functional properties of the IFM
Differences in troponin-tropomyosin interactions likely play critical roles in establishing and maintaining the specialized contractile characteristics of different muscles. For example, Drosophila asynchronous IFM displays a prominent stretch activation response, where stretch of the calcium-activated muscles triggers a delayed increase of force [43,44]. While this phenomenon is found in vertebrate striated muscles [45,46], in asynchronous IFM it is highly developed and is necessary for the oscillatory contractions required to power flight. An increased mass lying along Drosophila IFM tropomyosin may increase the stiffness of the regulatory strands, which could increase the cooperative unit of muscle activation well beyond the ~1.5 – 3 regulatory units proposed for vertebrates [5,6,47]. The greater stiffness may be instrumental in facilitating a minimum number of initially attached, stretched-distorted cross-bridges in further displacing extremely long, rigid segments of tropomyosin [48]. An explosive positive feedback mechanism would result and promote the rapid exposure and complete opening of myosin binding sites along distant thin filament regions. Thus, the greater regulatory strand density along IFM thin filaments may be crucial to the highly developed stretch activation response required for these specialized muscles to produce the oscillatory work necessary for flight. Such a role of TnT extensions in other arthropods remains uncertain. Further biochemical and physiological experimentation is required to determine if both Drosophila IFM and tarantula leg muscles share similar, enhanced cooperative activation and relaxation characteristics potentially attributed to the increased regulatory strand densities identified here.
Conclusions
Here we present the first structural evidence regarding the likely orientation of negatively charged extensions of TnT molecules within the troponin-tropomyosin regulatory complex. Such extensions are commonly found among invertebrates [49]. By performing cross-species comparisons of tropomyosin regulatory strand densities we confirmed significantly wider strands on arthropod thin filaments relative to those from vertebrates, although the arthropods’ strands did not significantly differ from each other. Drosophila indirect flight and tarantula femur muscle TnT isoforms contain Glu-rich C-terminal stretches that likely align and extend along tropomyosin and, consequently, add excess density discernable by electron microscopy and 3D reconstruction. In vertebrate muscle, TnT may contribute to regulating actomyosin ATPase activity and to establishing cooperative activation of the thin filament [1] in a Ca2+ sensitive manner [19,20]. Drosophila TnT possesses Ca2+-binding properties proposed to occur through its polyglutamic tail [24]. Thus, the tail’s extended orientation, optimally positioned along tropomyosin, suggests that in arthropods Ca2+ binding to TnT could substantially contribute to Ca2+ activation of the muscle by directly influencing the regulatory strands’ associations with actin and hence the occlusion of underlying myosin binding sites.
Acknowledgments
The authors thank S.I. Bernstein (San Diego State University) for critical reading of and helpful discussions regarding the manuscript. This work was funded by NIH grants AR034711 to RC and R01-HL36153 to WL.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gordon AM, Homsher E, Regnier M. Regulation of contraction in striated muscle. Physiol Rev. 2000;80:853–924. doi: 10.1152/physrev.2000.80.2.853. [DOI] [PubMed] [Google Scholar]
- 2.Brown JH, Cohen C. Regulation of muscle contraction by tropomyosin and troponin: how structure illuminates function. Adv Protein Chem. 2005;71:121–59. doi: 10.1016/S0065-3233(04)71004-9. [DOI] [PubMed] [Google Scholar]
- 3.Lehman W, Craig R. Tropomyosin and the steric mechanism of muscle regulation. Adv Exp Med Biol. 2008;644:95–109. doi: 10.1007/978-0-387-85766-4_8. [DOI] [PubMed] [Google Scholar]
- 4.McKillop DF, Geeves MA. Regulation of the interaction between actin and myosin subfragment 1: evidence for three states of the thin filament. Biophys J. 1993;65:693–701. doi: 10.1016/S0006-3495(93)81110-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vibert P, Craig R, Lehman W. Steric-model for activation of muscle thin filaments. J Mol Biol. 1997;266:8–14. doi: 10.1006/jmbi.1996.0800. [DOI] [PubMed] [Google Scholar]
- 6.Poole KJ, Lorenz M, Evans G, Rosenbaum G, Pirani A, Craig R, Tobacman LS, Lehman W, Holmes KC. A comparison of muscle thin filament models obtained from electron microscopy reconstructions and low-angle X-ray fibre diagrams from non-overlap muscle. J Struct Biol. 2006;155:273–84. doi: 10.1016/j.jsb.2006.02.020. [DOI] [PubMed] [Google Scholar]
- 7.Moore PB, Huxley HE, DeRosier DJ. Three-dimensional reconstruction of F-actin, thin filaments and decorated thin filaments. J Mol Biol. 1970;50:279–95. doi: 10.1016/0022-2836(70)90192-0. [DOI] [PubMed] [Google Scholar]
- 8.Phillips GN, Jr, Lattman EE, Cummins P, Lee KY, Cohen C. Crystal structure and molecular interactions of tropomyosin. Nature. 1979;278:413–7. doi: 10.1038/278413a0. [DOI] [PubMed] [Google Scholar]
- 9.Perry SV. Vertebrate tropomyosin: distribution, properties and function. J Muscle Res Cell Motil. 2001;22:5–49. doi: 10.1023/a:1010303732441. [DOI] [PubMed] [Google Scholar]
- 10.McLachlan AD, Stewart M. The 14-fold periodicity in alpha-tropomyosin and the interaction with actin. J Mol Biol. 1976;103:271–98. doi: 10.1016/0022-2836(76)90313-2. [DOI] [PubMed] [Google Scholar]
- 11.Greenfield NJ, Huang YJ, Swapna GV, Bhattacharya A, Rapp B, Singh A, Montelione GT, Hitchcock-DeGregori SE. Solution NMR structure of the junction between tropomyosin molecules: implications for actin binding and regulation. J Mol Biol. 2006;364:80–96. doi: 10.1016/j.jmb.2006.08.033. [DOI] [PubMed] [Google Scholar]
- 12.Greaser ML, Gergely J. Reconstitution of troponin activity from three protein components. J Biol Chem. 1971;246:4226–33. [PubMed] [Google Scholar]
- 13.Flicker PF, Phillips GN, Jr, Cohen C. Troponin and its interactions with tropomyosin. An electron microscope study. J Mol Biol. 1982;162:495–501. doi: 10.1016/0022-2836(82)90540-x. [DOI] [PubMed] [Google Scholar]
- 14.White SP, Cohen C, Phillips GN., Jr Structure of co-crystals of tropomyosin and troponin. Nature. 1987;325:826–8. doi: 10.1038/325826a0. [DOI] [PubMed] [Google Scholar]
- 15.Ohtsuki I, Nagano K. Molecular arrangement of troponin-tropomyosin in the thin filament. Adv Biophys. 1982;15:93–130. doi: 10.1016/0065-227x(82)90006-5. [DOI] [PubMed] [Google Scholar]
- 16.Potter JD, Gergely J. Troponin, tropomyosin, and actin interactions in the Ca2+ regulation of muscle contraction. Biochemistry. 1974;13:2697–703. doi: 10.1021/bi00710a007. [DOI] [PubMed] [Google Scholar]
- 17.Takeda S, Yamashita A, Maeda K, Maeda Y. Structure of the core domain of human cardiac troponin in the Ca(2+)-saturated form. Nature. 2003;424:35–41. doi: 10.1038/nature01780. [DOI] [PubMed] [Google Scholar]
- 18.Vinogradova MV, Stone DB, Malanina GG, Karatzaferi C, Cooke R, Mendelson RA, Fletterick RJ. Ca(2+)-regulated structural changes in troponin. Proc Natl Acad Sci U S A. 2005;102:5038–43. doi: 10.1073/pnas.0408882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dahiya R, Butters CA, Tobacman LS. Equilibrium linkage analysis of cardiac thin filament assembly. Implications for the regulation of muscle contraction. J Biol Chem. 1994;269:29457–61. [PubMed] [Google Scholar]
- 20.Potter JD, Sheng Z, Pan BS, Zhao J. A direct regulatory role for troponin T and a dual role for troponin C in the Ca2+ regulation of muscle contraction. J Biol Chem. 1995;270:2557–62. doi: 10.1074/jbc.270.6.2557. [DOI] [PubMed] [Google Scholar]
- 21.Bernstein SI, O'Donnell PT, Cripps RM. Molecular genetic analysis of muscle development, structure, and function in Drosophila. Int Rev Cytol. 1993;143:63–152. doi: 10.1016/s0074-7696(08)61874-4. [DOI] [PubMed] [Google Scholar]
- 22.Zhu J, Sun Y, Zhao FQ, Yu J, Craig R, Hu S. Analysis of tarantula skeletal muscle protein sequences and identification of transcriptional isoforms. BMC Genomics. 2009;10:117. doi: 10.1186/1471-2164-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fyrberg E, Fyrberg CC, Beall C, Saville DL. Drosophila melanogaster troponin-T mutations engender three distinct syndromes of myofibrillar abnormalities. J Mol Biol. 1990;216:657–75. doi: 10.1016/0022-2836(90)90390-8. [DOI] [PubMed] [Google Scholar]
- 24.Domingo A, Gonzalez-Jurado J, Maroto M, Diaz C, Vinos J, Carrasco C, Cervera M, Marco R. Troponin-T is a calcium-binding protein in insect muscle: in vivo phosphorylation, muscle-specific isoforms and developmental profile in Drosophila melanogaster. J Muscle Res Cell Motil. 1998;19:393–403. doi: 10.1023/a:1005349704790. [DOI] [PubMed] [Google Scholar]
- 25.Barbas JA, Galceran J, Krah-Jentgens I, de la Pompa JL, Canal I, Pongs O, Ferrus A. Troponin I is encoded in the haplolethal region of the Shaker gene complex of Drosophila. Genes Dev. 1991;5:132–40. doi: 10.1101/gad.5.1.132. [DOI] [PubMed] [Google Scholar]
- 26.Barbas JA, Galceran J, Torroja L, Prado A, Ferrus A. Abnormal muscle development in the heldup3 mutant of Drosophila melanogaster is caused by a splicing defect affecting selected troponin I isoforms. Mol Cell Biol. 1993;13:1433–9. doi: 10.1128/mcb.13.3.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mateos J, Herranz R, Domingo A, Sparrow J, Marco R. The structural role of high molecular weight tropomyosins in dipteran indirect flight muscle and the effect of phosphorylation. J Muscle Res Cell Motil. 2006;27:189–201. doi: 10.1007/s10974-005-9044-3. [DOI] [PubMed] [Google Scholar]
- 28.Newman R, Butcher GW, Bullard B, Leonard KR. A method for determining the periodicity of a troponin component in isolated insect flight muscle thin filaments by gold/Fab labelling. J Cell Sci. 1992;101( Pt 3):503–8. doi: 10.1242/jcs.101.3.503. [DOI] [PubMed] [Google Scholar]
- 29.Reedy MC, Reedy MK, Leonard KR, Bullard B. Gold/Fab immuno electron microscopy localization of troponin H and troponin T in Lethocerus flight muscle. J Mol Biol. 1994;239:52–67. doi: 10.1006/jmbi.1994.1350. [DOI] [PubMed] [Google Scholar]
- 30.Lehman W, Craig R, Vibert P. Ca(2+)-induced tropomyosin movement in Limulus thin filaments revealed by three-dimensional reconstruction. Nature. 1994;368:65–7. doi: 10.1038/368065a0. [DOI] [PubMed] [Google Scholar]
- 31.Cammarato A, Hatch V, Saide J, Craig R, Sparrow JC, Tobacman LS, Lehman W. Drosophila muscle regulation characterized by electron microscopy and three-dimensional reconstruction of thin filament mutants. Biophys J. 2004;86:1618–24. doi: 10.1016/S0006-3495(04)74229-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lehman W, Galinska-Rakoczy A, Hatch V, Tobacman LS, Craig R. Structural basis for the activation of muscle contraction by troponin and tropomyosin. J Mol Biol. 2009;388:673–81. doi: 10.1016/j.jmb.2009.03.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Craig R, Lehman W. The ultrastructural basis of actin filament regulation. Results Probl Cell Differ. 2002;36:149–69. doi: 10.1007/978-3-540-46558-4_12. [DOI] [PubMed] [Google Scholar]
- 34.Craig R, Lehman W. Crossbridge and tropomyosin positions observed in native, interacting thick and thin filaments. J Mol Biol. 2001;311:1027–36. doi: 10.1006/jmbi.2001.4897. [DOI] [PubMed] [Google Scholar]
- 35.Lehman W, Vibert P, Uman P, Craig R. Steric-blocking by tropomyosin visualized in relaxed vertebrate muscle thin filaments. J Mol Biol. 1995;251:191–6. doi: 10.1006/jmbi.1995.0425. [DOI] [PubMed] [Google Scholar]
- 36.Vibert P, Craig R, Lehman W. Three-dimensional reconstruction of caldesmon-containing smooth muscle thin filaments. J Cell Biol. 1993;123:313–21. doi: 10.1083/jcb.123.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Moody C, Lehman W, Craig R. Caldesmon and the structure of smooth muscle thin filaments: electron microscopy of isolated thin filaments. J Muscle Res Cell Motil. 1990;11:176–85. doi: 10.1007/BF01766496. [DOI] [PubMed] [Google Scholar]
- 38.Owen CH, Morgan DG, DeRosier DJ. Image analysis of helical objects: the Brandeis Helical Package. J Struct Biol. 1996;116:167–75. doi: 10.1006/jsbi.1996.0027. [DOI] [PubMed] [Google Scholar]
- 39.Trachtenberg S, DeRosier DJ. Three-dimensional structure of the frozen-hydrated flagellar filament. The left-handed filament of Salmonella typhimurium. J Mol Biol. 1987;195:581–601. doi: 10.1016/0022-2836(87)90184-7. [DOI] [PubMed] [Google Scholar]
- 40.Milligan RA, Flicker PF. Structural relationships of actin, myosin, and tropomyosin revealed by cryo-electron microscopy. J Cell Biol. 1987;105:29–39. doi: 10.1083/jcb.105.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Fyrberg C, Parker H, Hutchison B, Fyrberg E. Drosophila melanogaster genes encoding three troponin-C isoforms and a calmodulin-related protein. Biochem Genet. 1994;32:119–35. doi: 10.1007/BF00554420. [DOI] [PubMed] [Google Scholar]
- 42.Qiu F, Lakey A, Agianian B, Hutchings A, Butcher GW, Labeit S, Leonard K, Bullard B. Troponin C in different insect muscle types: identification of two isoforms in Lethocerus, Drosophila and Anopheles that are specific to asynchronous flight muscle in the adult insect. Biochem J. 2003;371:811–21. doi: 10.1042/BJ20021814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Abbott RH, Steiger GJ. Temperature and amplitude dependence of tension transients in glycerinated skeletal and insect fibrillar muscle. J Physiol. 1977;266:13–42. doi: 10.1113/jphysiol.1977.sp011754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pringle JW. Stretch-activation of muscle: function and mechanism. Proc R Soc B. 1978;201:107–130. doi: 10.1098/rspb.1978.0035. [DOI] [PubMed] [Google Scholar]
- 45.Steiger GJ. Stretch-activation and tension transients in cardiac, skeletal and insect flight muscle. In: Tregear RT, editor. Insect Flight Muscle. Elsevier; Amsterdam: 1977. pp. 221–268. [Google Scholar]
- 46.Vemuri R, Lankford EB, Poetter K, Hassanzadeh S, Takeda K, Yu ZX, Ferrans VJ, Epstein ND. The stretch-activation response may be critical to the proper functioning of the mammalian heart. Proc Natl Acad Sci U S A. 1999;96:1048–53. doi: 10.1073/pnas.96.3.1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lehrer SS, Geeves MA. The muscle thin filament as a classical cooperative/allosteric regulatory system. J Mol Biol. 1998;277:1081–9. doi: 10.1006/jmbi.1998.1654. [DOI] [PubMed] [Google Scholar]
- 48.Linari M, Reedy MK, Reedy MC, Lombardi V, Piazzesi G. Ca-activation and stretch-activation in insect flight muscle. Biophys J. 2004;87:1101–11. doi: 10.1529/biophysj.103.037374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Benoist P, Mas JA, Marco R, Cervera M. Differential muscle-type expression of the Drosophila troponin T gene. A 3-base pair microexon is involved in visceral and adult hypodermic muscle specification. J Biol Chem. 1998;273:7538–46. doi: 10.1074/jbc.273.13.7538. [DOI] [PubMed] [Google Scholar]