Abstract
The NF-κB family of transcription factors plays an important role in determining cell survival during immune, inflammatory, and stress responses. NF-kB activity is frequently deregulated in human cancers and is implicated in the resistance of tumor cells to diverse anticancer agents. We studied the effects of novel analogs of precursors of the natural product simplactone (A) on the activity of IkB kinase and NF-kB. Screening of six compounds for the ability to inhibit TNF-induced NF-kB activity revealed that compound SK2009 was the most potent of these compounds in suppressing NF-kB activation in KBM-5 leukemic cells. Further characterization of SK2009 indicates that this newly synthesized molecule can suppress TNF-induced IκBα kinase activation and inhibit the expression of three NF-kB-dependent gene products, cyclin D1, Bcl-2, and VEGF, in these cells.
Keywords: Novel chemical entities, TNF, NF-kB, IKK, KBM-5 cells, simplactones
1. Introduction
The NF-κB family of heterodimeric transcription factors (Rel proteins) plays an important role in determining cell survival during immune, inflammatory, and stress responses. NF-κB/Rel proteins share a highly conserved N-terminal Rel homology domain (RHD) that is responsible for DNA binding and dimerization. Mammals express five Rel proteins that belong to two classes.1 Members of one group (RelA, c-Rel, and RelB) are synthesized as mature proteins, while the other group (encoded by NFkB1 and NFkB2) includes precursor proteins (p105 and p100, respectively) that undergo proteolysis to yield their mature products (the p50 and p52 NF-κB proteins). NF-kB typically exists as a heterodimeric complex composed of the Rel family proteins p50 and p65. It usually resides in the cytoplasm in an inactive form as a result of its association with its inhibitor IkB. A number of extracellular signals can lead to NF-kB activation through the phosphorylation and degradation of IkB. The liberated NF-kB is then translocated to the nucleus, where it binds to specific promoters and regulates target gene expression. In normal cells, NF-kB is strictly regulated, whereas it is often constitutively activated to high levels in cancer cells.2 More importantly, NF-kB activation in cancer cells has been shown in many studies to be one of the major causes of resistance to chemotherapy.3,4
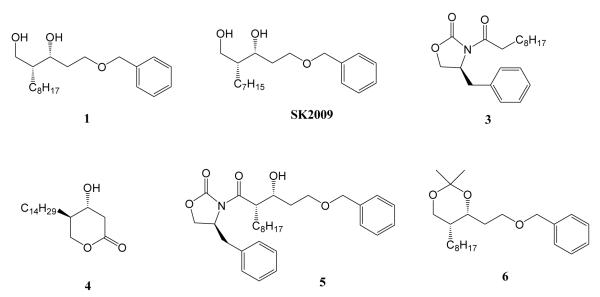
Since NF-kB represents an important and attractive therapeutic target for treating a variety of diseases, a great deal of attention has been focused on the identification of compounds that selectively interfere with this pathway. Recently, a large number of natural products have been evaluated as possible inhibitors of NF-kB, including lignins, diterpenes, triterpenes, and polyphenols5,6. We have previously described the synthesis of simplactone and it’s analog and evaluated their activity against anticancer cell lines6. We have synthesized six new compounds that are analogs of precursors of the natural product simplactone (Figure 1).8,9 In the current study, we examined the ability of these compounds to suppress the TNFα-induced NF-kB activation, and identified SK2009 as an effective inhibitor of this signaling pathway.
Figure 1.
Chemical Structures of the compounds
2. Results
2.1 Chemistry
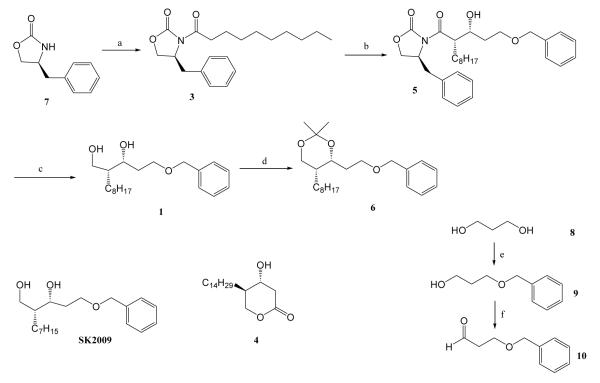
Our synthetic route for all the compounds involved Evans asymmetric aldol methodology (Scheme 1) to set up the stereochemistry of the two contiguous stereogenic centers.9 Compound 7, (S)-4-Benzyl-2-oxazolidinone, was coupled with the indicated acids in the presence of pivaloyl chloride and triethyl amine to yield 3. Treatment of the boron enolate derived from compound 3 with aldehyde 10 provided Compound 5 in 91% yield, which was converted to diol 1 after removal of the oxazolidinone auxiliary with aqueous NaBH4. Diol 1 then underwent acetonide protection using 2,2-dimethoxy propane (2,2-DMP) to generate compound 6. SK2009 was synthesized using a procedure similar to that used to prepare compound 1, substituting the appropriate acid. To synthesize aldehyde 10, one of the hydroxyl functionalities of propanediol 8 was protected as benzyl ether 9, and the other hydroxyl functionality was oxidized using Swern oxidation conditions. Compound 4 was synthesized using the methodology we have previously reported.11
Scheme 1.
Reagents and conditions: (a) CH3(CH2)8COOH(2), pivaloyl chloride, Et3N, LiCl, THF; (b) Bu2BOTf, Et3N, BnO(CH2)2CHO(10); (c) Aq. NaBH4, THF; (d) 2,2-DMP, CH2Cl2, PTSA; (e) 8, NaH, DMF, BnBr; (f) (COCl)2, DMSO, CH2Cl2
2.2. Biology
2.2.1. Cell lines and reagents
The cell line KBM-5 (human chronic myeloid leukemia) was obtained from the American Type Culture Collection and was cultured in Iscove’s modified Dulbecco’s medium with 15% FBS. Culture media were supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. Bacteria-derived human recombinant human TNFα, purified to homogeneity with a specific activity of 5 × 107 U/mg, was kindly provided by Genentech (South San Francisco, CA, USA). Antibodies against cyclin D1, Bcl-2 and VEGF were obtained from Santa Cruz Biotechnology (Santa Cruz, CA, USA). Anti-IKK-α, and anti-IKK-β, antibodies were kindly provided by Imgenex (San Diego, CA, USA).
2.2.2. Electrophoretic mobility shift assays
For all invitro studies 50mM stock of all the compounds were made in DMSO with subsequent dilution in cell culture media. To assess NF-κB activation, nuclear extracts of KBM-5 cells (1×106/mL) either treated or untreated with TNFα were prepared and electrophoretic mobility shift assay (EMSAs) were performed as described previously.12,13 In brief, nuclear extracts were incubated with 32P-end-labeled 45-mer double-stranded oligonucleotide (15 μg protein with 16 fmol DNA) from the HIV long terminal repeat, 5 -TTGTTACAA GGGACTTTC CGCTG GGGACTTTC CAGGGAGGCGTGG- 3 (boldface indicates NF-κB binding sites), for 30 min at 37°C. The DNA-protein complex formed was separated from free oligonucleotide on 6.6% native polyacrylamide gels. The dried gels were visualized with a Storm 820 imaging system, and radioactive bands were quantified using Imagequant software Imagequant software version 3.3 (Molecular Dynamics, Piscataway, NJ).
2.2.3. Western blot analysis
Western blot analysis of KBM-5 cell lysates was performed as described previously.14 30-μg protein samples were resolved on SDS-PAGE and probed with specific antibodies for cyclin D1, Bcl-2, and VEGF (1:1000 dilutions). The blots were washed, exposed to HRP-conjugated secondary antibody (1:3000 dilutions) for 2 h, and finally detected by enhanced chemiluminescence (ECL) reagent (Amersham Pharmacia Biotechnology). The bands were quantified with a Personal Densitometer Scan v1.30 using Imagequant software version 3.3 (Molecular Dynamics, Piscataway, NJ).
2.2.4. Immunoprecipitation assays
The IKK complex from whole-cell extracts of KBM-5 cells was precipitated with anti-IKK-α antibody and then incubated with protein A/G-agarose beads (Pierce Chemical, Rockford, IL). After 2 h, the beads were washed with lysis buffer and resuspended in a kinase assay mixture containing 50 mM HEPES (pH 7.4), 20 mM MgCl2, 2 mM dithiothreitol, 20 μCi (0.74 MBq) –[32P]ATP, 10 mM unlabeled ATP, and 2 μg of substrate (GST-IκBα, amino acids 1-54) and incubated at 30°C for 30 min. The reaction was terminated by boiling in SDS sample buffer for 5 min. Finally, the protein was resolved on 10% SDS-PAGE, the gels were dried, and the radioactive bands were visualized with a Storm 820 system. To determine the total amounts of IKK-α and IKK-β in each sample, 30 μg of whole-cell proteins was resolved on 10% SDS-PAGE, electro blotted to a nitrocellulose membrane, and then reacted with either anti–IKK-α or anti–IKK-β antibody.
2.2.5. Cytotoxicity assay
Cytotoxicity was assayed by the modified tetrazolium salt 3-(4-5-dimethylthiozol-2-yl)2-5-diphenyl-tetrazolium bromide (MTT) assay (Mossman, 1983). Briefly, KBM-5 cells (5000 per well) were incubated in triplicate in a 96-well plate in the presence or absence of compounds 1-6 for 72 h at 37 °C. Thereafter, 20 μl MTT solution (5 mg/ml in PBS) was added to each well. After a 2-h incubation at 37 °C, 0.1 mL of extraction buffer (20% SDS, 50% dimethylformamide) was added; incubation was continued overnight at 37 °C; and then the optical density (OD) at 570 nm was measured by means of a 96-well multiscanner auto reader (Dynatech MR 5000, Chantilly, VA).
3. Discusion
The purpose of the present study was to synthesize analogs of precursors of simplactone and evaluate their ability to modulate the NF-κB signaling pathway, NF-κB-regulated gene activity, and NF-κB-mediated cellular responses. We chose KBM-5 cells for our experiments because these cells are known to express both types of TNF receptors, and the TNFα-induced NF-κB activation pathway is well characterized in this cell line.
3.1. All six of the synthesized compounds suppress TNF-induced NF-κB activation
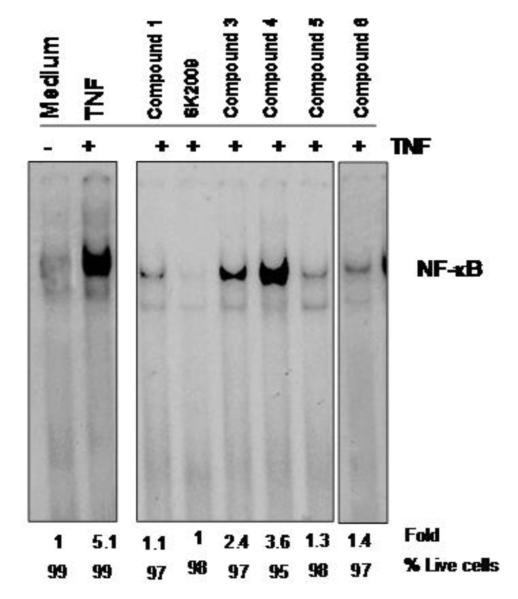
We initially screened all six compounds (Figure 1) for the ability to inhibit TNF-induced NF-κB activation. All the six compounds were able to inhibit the activation of NF-κB without adversely affecting cell viability (viability remained >=95%). SK2009 was clearly the most potent of these compounds in terms of suppressing NF-κB activation (inhibiting the activity by approximately 5-fold; Figure 2).
Figure 2.
Effect of compounds on TNF-induced NF-κB activation. KBM-5 cells (1×106 cells/mL) were incubated with each of the compounds at 100 μM for 12 h and then treated with 0.1 nM TNF for 30 min. Nuclear extracts were prepared and assayed for NF-κB activation by EMSA. Cell viability was measured by trypan blue assay. Data is the average of 3 independent experiments.
3.2. SK2009 inhibits TNF-induced NF-κB activation in a dose-dependent manner
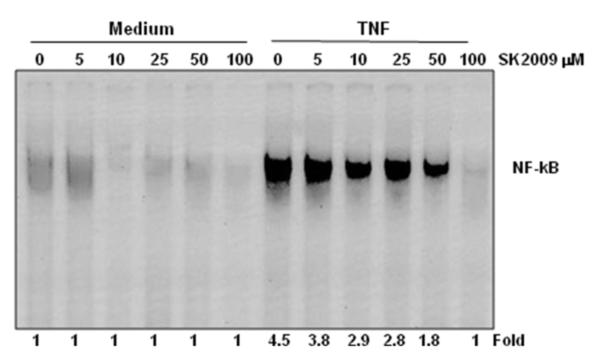
We determined the minimum dose of SK2009 that was required to inhibit TNF-induced NF-κB activation. For this purpose, KBM-5 cells were treated with SK2009 at concentrations of 5-100 μM and then exposed to TNFα to activate NF-κB. SK2009 alone had no effect on basal NF-κB activity, but it was able to suppress TNFα-induced NF-κB activation in a dose-dependent manner, with maximal inhibition (4.5-fold) occurring at 100 μM (Figure 3).
Figure 3.
Dose-dependent effect of SK2009 on TNF-induced NF-κB activation. KBM-5 cells (1×106 cells/mL) were incubated with the indicated concentrations of SK2009 for 12 h and treated with 0.1 nM TNF or medium alone (−TNF) for 30 min. nuclear extracts were prepared and assayed for NF-κB activation by EMSA. Data is the average of 3 independent experiments.
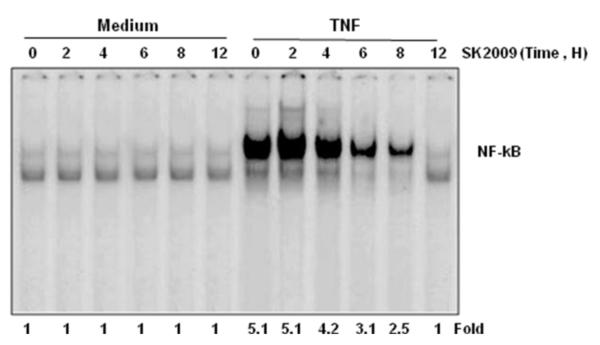
3.3. SK2009 inhibits TNF-induced NF-κB activation in a time-dependent manner
To investigate the duration of exposure required for inhibition of NF-κB by SK2009, KBM-5 cells were exposed to SK2009 at 100 μM for different time intervals prior to treatment with TNF. EMSA of nuclear extracts prepared from TNF-treated and untreated cells indicated that SK2009 inhibited TNF-mediated NF-κB activation in a time-dependent manner, with maximal inhibition occurring after 12h of incubation with SK2009 (Figure 4).
Figure 4.
Time-dependent effect of SK2009 on TNF-induced activation of NF-κB. KBM-5 cells (1×106 cells/mL) were incubated with SK2009 at 100 μM for the indicated times and then treated with 0.1 nM TNF for 30 min. Nuclear extracts of the cells were then prepared and assayed for NF-kB activation using EMSA. Data is the average of 3 independent experiments.
3.4. SK2009 does not directly interact with the NF-κB p65 subunit
To investigate whether SK2009 directly interacts with the RelA/p65 subunit of NF-κB, nuclear extracts isolated from TNF-treated and untreated KBM-5 cells were exposed to SK2009 at various concentrations, and their binding to DNA was examined by EMSA. As shown in Figure 5, SK2009 did not interfere with the binding of p65 to DNA.
Figure 5.
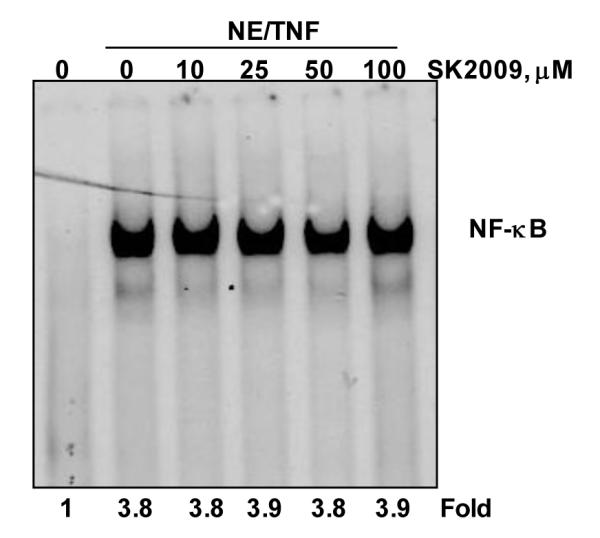
SK2009 did not inhibit the direct binding of NF-κB to DNA. Nuclear extracts (NE) were prepared from untreated KBM-5 cells or cells treated with 0.1 nM TNF for 30 min, incubated for 30 min with the indicated concentrations of SK2009, and assayed for DNA binding by EMSA. Data is the average of 3 independent experiments.
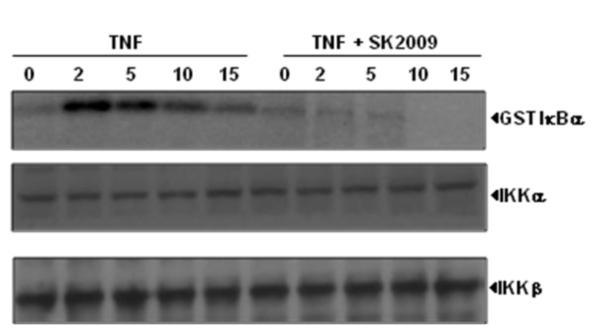
3.5. SK2009 suppresses TNF-induced IκBα kinase (IKK) activation in vivo
Activation of IKK, a multi-subunit complex comprising the catalytic subunits IKK-α and IKK-β, is essential for the TNF-induced phosphorylation and degradation of IκBα. We investigated whether SK2009 inhibits TNF-induced activation of IKK in KBM-5 cells. The analysis of kinase activity of IKKα immunoprecipitates from whole cell lysates indicated that TNF activated IKK activity in a time-dependent manner, and that SK2009 inhibited the kinase activity of IkBα kinase (Figure 6). Neither TNF nor SK2009 had any effect on the protein levels of either IKK-α or IKK-β.
Figure 6.
Effect of SK2009 on the activation of IκBα kinase (IKK) by TNF. KBM-5 cells (5×106/mL) were preincubated with SK2009 at 100 μM for 12 h, then treated with 1 nM TNF for the indicated times. Whole-cell lysates were prepared and immunoprecipitated with an anti-IKK-α antibody, then analyzed by kinase assay. To examine the effect of SK2009 on the level of expression of IKK proteins, whole-cell extracts were fractionated on SDS-PAGE and examined by Western blot analysis using anti-IKK-α and anti-IKK-β antibodies. Data is the average of 3 independent experiments.
3.6. SK2009 has no effect on TNF-induced IKK activation in vitro
To determine whether SK2009 suppresses IKK activity directly by binding to IKK, the TNF treated and untreated cell extracts were immunoprecipitated with anti-IKK-α antibody and these immune complexes were incubated with different concentrations of SK2009. Examination of IKK activity in IKK-α precipitated immune complex showed that SK2009 did not directly inhibit the activity of IKK in vitro (Figure 7).
Figure 7.
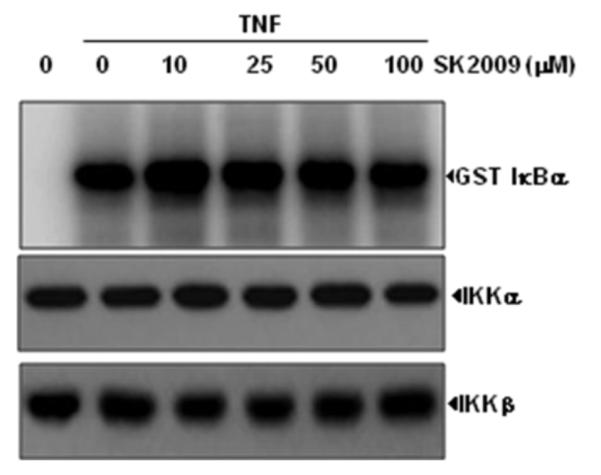
Lack of a direct effect of SK2009 on IKK activation induced by TNF. Whole-cell extracts were prepared from KBM-5 cells that had been treated with 1nM TNF and immunoprecipitated with anti-IKK-α antibody. Kinase assays of the immune complexes were performed in the absence or presence of the indicated concentrations of SK2009. Data is the average of 3 independent experiments.
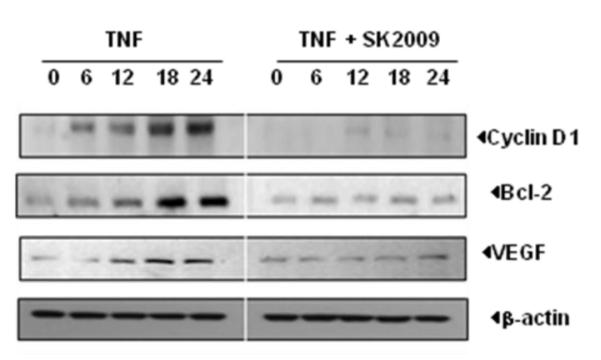
3.7. SK2009 suppresses the expression of NF-κB-dependent gene products
Several studies have linked activation of NF-κB pathway with tumor cell survival. Because SK2009 inhibits TNF-induced activation of NF-κB, we examined whether it inhibits TNF-induced expression of NF-κB linked genes. KBM-5 cells were cultured in the presence SK2009, and then co-incubated with TNF for the indicated time points. Immunoblot analysis of whole-cell extracts showed that SK2009 suppressed the expression of the NF-κB-dependent proteins cyclin D1, Bcl-2, and VEGF (Figure 8).
Figure 8.
SK2009 suppresses the expression of TNF-induced cyclin D1, Bcl-2, and VEGF. KBM-5 cells (1×106/mL) were co- incubated with SK2009 at 100 μM concentration along with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and fractionated using 10%vSDS-PAGE, and electro-transferred to a nitrocellulose membrane and analyzed by Western blotting using antibodies specific for the individual proteins. Data is the average of 3 independent experiments.
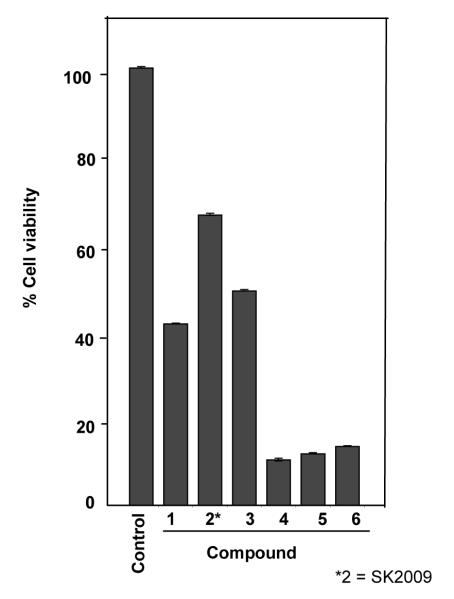
3.8. SK2009 suppresses the survival of KBM-5 cells in culture
We evaluated the relative ability of the six compounds to inhibit the survival of the leukemic KBM-5 cells in culture. Compound 4 was found to be more potent in inhibiting the cell survival, and SK2009 had the least dramatic effect on cell survival (Figure 9).
Figure 9.
Effect of all the six compounds on cell survival. KBM 5 cells (5×103cells/well in a 96 well format) were incubated with 10 μM of each compound for 72 h, and cell viability was determined by MTT assay. Data is the average of 3 independent experiments.
Since the presence of a long chain of carbon atoms in a bioactive molecule might play a role in the activity profile of the molecule, we synthesized six analogs of the natural product, simplactone, with different carbon chain length and investigated their role in TNF-induced NF-κB activation as well as TNF-mediated cellular responses. The TNF mediated NF-kB activation pathways are relatively well established so we decided to focus this signaling and investigated effect of simplactone analogs. Of the six analogs of simplactone, SK2009 was the most effective inhibitor of TNF induced NF-κB activation. We found that SK 2009 inhibits the NF-kB activation induced by TNF in a dose and time dependent manner.
The phosphorylation of p65 subunit is necessary for the nuclear translocation and transcriptional activity of NF-kB. IKK is the kinase that phosphorylate p65 subunit. Thus we decided to investigate the effect of SK2009 on the kinase activity of IKK alpha subunit. We found that in SK 2009 pretreated cells TNF failed to stimulate the kinase activity of IKK.
During the course of our investigation we found that various gene products which are regulated by FkB were also down regulated by SK 2009. These proteins include cyclin D1 (a cell cycle regulator involved in cellular proliferation), Bcl-2 (an anti-apoptotic protein) and VEGF (which regulates angiogenesis).
Constitutive activation of NF-κB in cancer cells has been reported to promote cell survival and protect tumor cells from TNFa or chemotherapeutic agents.15 However, we could not correlate the inhibition of NF-κB activation with the direct cellular cytotoxicity of the tested compounds. SK2009 has the maximum inhibition of NF-κB activation with least cytotoxic potential as compared to compound 4 which shows maximum cytotoxic potential. The inhibition of NF-κB activation of SK2009 is 100% as compared to compound 4 (30%). All the six synthetic precursor analogs of simplactone show biological activity as compared to their parent compound. Further modifications of SK2009 may yield analogs with improved biological activity and antitumor efficacy.
4. Experimental
4.1 General
All reagents and solvents were obtained from Aldrich. Anhydrous solvents were used as received. Reaction progress was monitored with analytical thin-layer chromatography (TLC) plates on 0.25 mm Merck F-254 silica gel glass plates. Visualization was achieved using UV illumination. 1H NMR spectra were obtained at 400 MHz on a Bruker Avance spectrometer and are reported in parts per million downfield relative to tetramethylsilane (TMS). EI-MS profiles were obtained using a Bruker Esquire 3000 plus. The HPLC method was set up on a Waters System equipped with a PDA detector 2996, using C18 column (5 μm, 250 × 4.6 mm) with water and acetonitrile as mobile phase and the flow rate of 1 mL/min. The wavelengths were obtained at 214 nm.
4.2 : General Procedure for the preparation of compounds 1-10
(S)-4-benzyl-3-decanoyloxazolidin-2-one (3)
To a solution of decanoic acid (0.2 g, 1.0 mmol) in THF (5 mL) at −20°C was added triethylamine (0.18 mL, 1.3 mmol), LiCl (catalytic amount) and pivaloyl chloride (0.13 mL, 1.1 mmol). The reaction mixture was stirred at −78°C for 1 h and the resulting pivaloyl anhydride was used without purification. To this reaction mixture a solution of oxazolidinone 7 (0.212 g, 1.2 mmol) in THF (5 mL) was added at −78°C. The reaction mixture was stirred at this temperature for 1 h, quenched with satd. NH4Cl (2 mL), and washed with Et2O (2×10 mL). The combined organic phases were washed with satd. NaHCO3 (5 mL), brine (5 mL), dried (MgSO4) and concentrated. Purification by flash chromatography (SiO2, EtOAc/hexane 1:4) gave 0.276 g (78%) of 3 as a yellow oil; 1H NMR (400 MHz, CDCl3): 7.23-7.42 (m, 5H), 4.67-4.73 (m, 1H), 4.17-4.24 (m, 2H), 3.34 (dd, 1H, J = 3.2, 13.2 MHz), 2.88-3.03 (m, 2H), 2.79 (ab q, 1H, J = 9.6, 13.2 MHz), 1.68-1.76 (m, 2H), 1.23-1.46 (m, 12 H), 0.91 (t, 3H, J = 6.4, 13.6 MHz); MS: 332 (M++1).
(S)-3-((S)-2-((R)-3-(benzyloxy)-1-hydroxypropyl)decanoyl)-4-benzyloxazolidin-2-one (5)
To a solution of oxazolidinone 3 (0.9 g, 3.6 mmol) in CH2Cl2 (10 mL) at 0°C, Dibutyl boron triflate (0.79 mL, 7.2 mmol) was added dropwise. The reaction mixture turned into yellowish slurry. It was stirred for 5 min at 0°C and then diisopropylethylamine (0.7 mL, 3.96 mmol) was added dropwise. The reaction mixture now turned deep reddish in color. Stirring was continued for another 20 min at 0°C. It was cooled to −78°C and aldehyde 10 (0.5 mL, 3.96 mmol) in CH2Cl2 (3 mL)was added dropwise, during which the yellow color appeared again. Then it was stirred at –78°C for 1 h, warmed to 0°C and quenched by the addition of cold saturated NH4Cl solution. It was then extracted with EtOAc (20 mL) and washed with brine (10mL), dried (Na2SO4) and concentrated in vacuo. Purification by column chromatography (SiO2, EtOAc/Hexanes 2:4) afforded 5 (12.25 g, 88%) as a colorless solid. 1H NMR (400 MHz, CDCl3): 7.23-7.40 (m, 10H), 4.65-4.72 (m, 1H), 4.53 (s, 2H), 4.11-4.19 (m, 5H), 3.66-3.73 (m, 2H), 3.30-3.37 (m, 1H), 3.15-3.19 (m, 1H), 2.69-2.75 (dd, 1H, J = 10.4, 13.2 MHz), 1.83-1.87(m, 2H), 1.26-1.40 (m, 12H), 0.88 (t, 3H, J = 6.8, 13.6 MHz); MS: 518 (M++Na), 497 (M++1).
(2R,3R)-5-(benzyloxy)-2-octylpentane-1,3-diol (1)
To a solution of compound 5 (1.0 g, 2 mmol) in THF (5 mL) was added sodium borohydride (0.1g, 2.4 mmol ) and 2 drops of water at 0°C and the reaction mixture was stirred for 1 hour at the same temperature. Reaction progress was monitored by TLC and quenched the reaction mixture with saturated NH4Cl solution (5 mL) and extracted with ethyl acetate twice (2 × 10 mL). Combined organic layers were washed with brine (20 mL), dried over Na2SO4 and concentrated in vacuo. Purification by column chromatography (SiO2, EtOAc/ Hexanes 6:4) afforded diol 1 as a pure thick syrup. 1H NMR (400 MHz, CDCl3): 7.28-7.39 (m, 5H), 4.55 (s, 2H), 4.07-4.15 (m, 1H), 3.64-3.83 (m, 5H), 3.07-3.11 (m, 1H), 1.97-2.03 (m, 2H), 1.58-1.65 (m, 2H), 1.22-1.33 (m, 10H), 0.90 (t, 3H, J = 6.4, 13.2 MHz)
MS: 322 (M++1).
(4R,5R)-4-(2-(benzyloxy)ethyl)-2,2-dimethyl-5-octyl-1,3-dioxane (6)
To a solution of compound 1 (0.7g, 2.2 mmol) in CH2Cl2 (10 mL) was added 2,2-dimethoxy propane (1.2 g, 11 mmol) and catalytic amount of p-Toluene-sulfonic acid and stirred the reaction mixture for 4 hours at ambient temperature. To the crude compound obtained after removal of the solvents was added water (10 mL) and extracted with ethyl acetate twice (2 × 30 mL). Combined organic layers were washed with brine (20 mL), dried over Na2SO4 and concentrated in vacuo. Purification by column chromatography (SiO2, EtOAc/ Hexanes 3:7) afforded 6 as a syrup. 1H NMR (400 MHz, CDCl3): 7.35-7.38 (m, 5H), 4.51 (d, 2H, J = 3.6 MHz), 4.17-4.22 (m, 1H), 3.95-4.01 (m, 1H), 3.77-3.81 (m, 1H), 3.51-3.58 (m, 2H), 1.80-1.90 (m, 1H), 1.60-1.76 (m, 2H), 1.44 (s, 3H), 1.38 (s, 3H), 1.25-1.35 (m, 14H), 0.90 (t, 3H, J = 6.8, 13.6 MHz); MS: 362 (M++1).
(2R,3R)-5-(benzyloxy)-2-heptylpentane-1,3-diol (SK2009)
SK2009 was synthesized based on the synthesis of 5 using corresponding acid CH3 (CH2)7COOH.
1H NMR (400 MHz, CDCl3): 7.30-7.40 (m, 5H), 4.56 (s, 2H), 4.11 (d, 1H, J = 10.8 MHz), 3.76-3.86 (m, 2H), 3.67-3.73 (m, 2H), 1.87-2.10 (m, 1H), 1.19-1.42 (m, 12H), 0.90 (t, 3H, J = 6.4, 13.2 MHz); MS: 309 (M++1).
(4R,5S)-tetrahydro-4-hydroxy-5-tetradecylpyran-2-one (4)
Compound 4 was synthesized using the reference 7.
1H NMR (400 MHz, CDCl3): 4.5 (dd, 1H, J = 4.4, 11.6 MHz), 3.94-4.0 (m, 2H), 2.90 (dd, 1H, J = 6.0, 17.6 MHz), 2.56 (dd, 1H, J = 6.0, 17.6 MHz), 1.81-1.97 (m, 2H), 1.54-1.66 (m, 2H), 1.25-1.31 (m, 24H), 0.90 (t, 3H, J = 6.8, 13.6 MHz); MS: 647 (dimmer), 313 (M++1).
3-Benzyloxy-1-propionaldehyde (10)
DMSO (1.6 mL, 22 mmol) was added to a solution of (COCl)2 (1.0 mL, 11 mmol) in CH2Cl2 (100 mL) at −78°C. The reaction mixture was stirred for 10 min at which time 3-benzyl-1-propyl alcohol (9, 1.66 g, 10.0 mmol) was added dropwise in CH2Cl2 (5 mL). After 15 min, triethylamine (7.0 mL, 50 mmol) was added, the solution was stirred for 5 min at which time the reaction mixture was warmed to room temperature. After stirring for 30 min the reaction was quenched with saturated NH4Cl (10 mL), and the aqueous phase was washed with CH2Cl2 (2×20 mL). The combined organic phase was washed with saturated NaHCO3 (2×10 mL), brine (10 mL), dried (Na2SO4) and concentrated. Purification by flash chromatography (EtOAc/hexane 1:4) gave aldehyde 10 as a clear oil (1.63 g, 99%); 1H NMR (CDCl3) δ 7.2–7.4 (m, 5H), 4.5 (s, 2H), 3.8–3.9 (t, 2H, J=6.1 MHz), 2.6–2.7 (m, 2H);
5. Conclusion
In summary, we report the synthesis and biological evaluation of novel promising candidates that are able to inhibit TNF-induced NF-κB activation in KBM-5 cells in a dose- and time-dependent manner. The results showed that SK2009 (branched aliphatic alcohol) which is a precursor of simplactone analog suppressed TNF-induced IκBα kinase (IKK) activation but no intrinsic activity on TNF-induced IκBα kinase (IKK) activation in in vitro experiments. Interestingly, SK2009 showed a potential suppression on the expression of TNF-induced cyclin D1, Bcl-2, and VEGF in KBM-5 cells. Detail studies of molecular modeling based drug design are underway of SK2009 in order to obtain improved activity at a lower dose.
6. Acknowledgements
We gratefully acknowledge the financial support of FAMRI grant (to S.R.K). Dr. Aggarwal is Ransom Horne, Jr., Professor of Cancer Research. This work was supported by a grant from the Clayton Foundation for Research (to B. B. A.).
Abbreviations
- TNF
Tumor Necrosis Factor
- NF-κB
nuclear factor-kappa B
- IKK
Ikappa B Kinase
- 2,2-DMP
2,2-dimethoxy propane
- NaBH4
Sodium Borohydride
- HEPES
4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
7. References
- 1.Karin M, Ben NY. Annu. Rev. Immunol. 2000;18:621–663. doi: 10.1146/annurev.immunol.18.1.621. [DOI] [PubMed] [Google Scholar]
- 2.Pacifico F, Leonardi A. Biochem Pharmacol. 2006;72:1142–1152. doi: 10.1016/j.bcp.2006.07.032. [DOI] [PubMed] [Google Scholar]
- 3.Wang CY, Cusack JC, Liu R, Baldwin AS. Nat Med. 1999;5:412–417. doi: 10.1038/7410. [DOI] [PubMed] [Google Scholar]
- 4.Katsman A, Umezawa K, Bonavida B. Drug Resist Update. 2007;10:1–12. doi: 10.1016/j.drup.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 5.De Guzman FS, Schmitz FJ. J. Nat. Prod. 1990;53:926–931. [Google Scholar]
- 6.McKenzie M, Carl Li, Kaufman P, Seymour EM, Kirakosyan A. Recent-Advances in Plant Biotechnology: Book Chapter 10.1007/978-1-4419-0194-1_11.
- 7.Cafieri F, Fattorusso E, Taglialatela-Scafati O, Di Rosa M, Ianaro I. Tetrahedron. 1999;55:13831–13840. [Google Scholar]
- 8.Evans DA, Batroli J, Shih TL. J. Am. Chem. Soc. 1981;103:2127–2129. [Google Scholar]
- 9.Anchoori RK, Sudhakar N, Venkateswara Rao B, Raghunandan N, Venkatesh A, Sarangapani M. Bioorganic & Medicinal Chemistry Letters. 2005;15:2085–2086. doi: 10.1016/j.bmcl.2005.02.059. [DOI] [PubMed] [Google Scholar]
- 10.Mosmann T. J Immunol Methods. 1983;65:55–63. doi: 10.1016/0022-1759(83)90303-4. [DOI] [PubMed] [Google Scholar]
- 11.Chaturvedi MM, Mukhopadhyay A, Aggarwal BB. Methods Enzymol. 2000;319:585–602. doi: 10.1016/s0076-6879(00)19055-x. [DOI] [PubMed] [Google Scholar]
- 12.Kunnumakkara AB, Nair AS, Ahn KS, Pandey MK, Yi Z, Liu M, Aggarwal BB. Blood. 2007;15:5112–5121. doi: 10.1182/blood-2007-01-067256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Aggarwal BB. Cancer Cell. 2004;6:203–8. doi: 10.1016/j.ccr.2004.09.003. [DOI] [PubMed] [Google Scholar]
- 14.Aggarwal S, Ichikawa H, Takada Y, Sandur SK, Shishodia S, Aggarwal BB. Mol Pharmacol. 2006;69:195–206. doi: 10.1124/mol.105.017400. [DOI] [PubMed] [Google Scholar]
- 15.Manna SK, Mukhopadhyay A, Aggarwal BB. J. Immunol. 2000;164:6509–19. doi: 10.4049/jimmunol.164.12.6509. [DOI] [PubMed] [Google Scholar]