Figure 3.
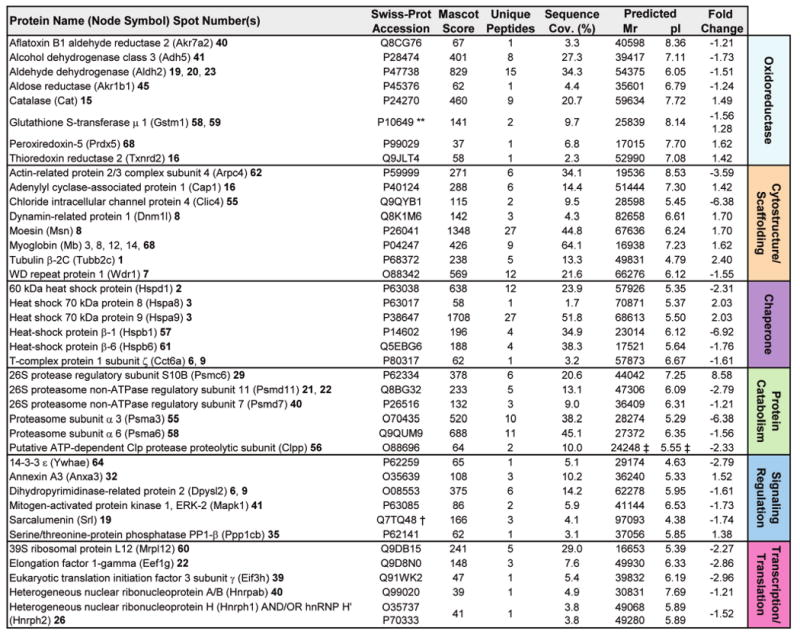
Secondary functions associated with KATP channel deficiency form an infrastructure supporting metabolism. Ontological categorization indicated that a secondary functional group consisting of the remaining identified proteins (39/102) comprises a number of functions supporting cellular metabolic activity. These proteins function as oxidoreductases, in cytostructure and scaffolding, as stress-related chaperones, or in protein catabolism, signaling regulation, or transcription and translation. Protein names are listed with their Swiss-Prot gene name (for Node Symbol) to locate them in the protein interaction network, and with their corresponding spot numbers from their 2-D gel positions. Protein accession number, Mascot score, number of unique identified peptides, % sequence cov. (coverage), predicted Mr and pI for each protein (following expected post-translational processing, for example, removal of a mitochondrial signal peptide), and fold change (KO versus WT) are indicated. For proteins detected in more than one spot, maximum score and number of unique peptides are reported. Fold change was calculated as described in experimental procedures, and for proteins detected in both increasing and decreasing spots (**), both values are indicated. Complete MS/MS data for all proteins is outlined in Supplementary Table 1 (Supporting Information). (**Contains both up- and down-regulated spots; † Node not detected for network analysis by Ingenuity Pathways; ‡ Mr/pI calculated after TargetP 1.1 prediction of mitochondrial localization and signal peptide cleavage site).
