Figure 5.
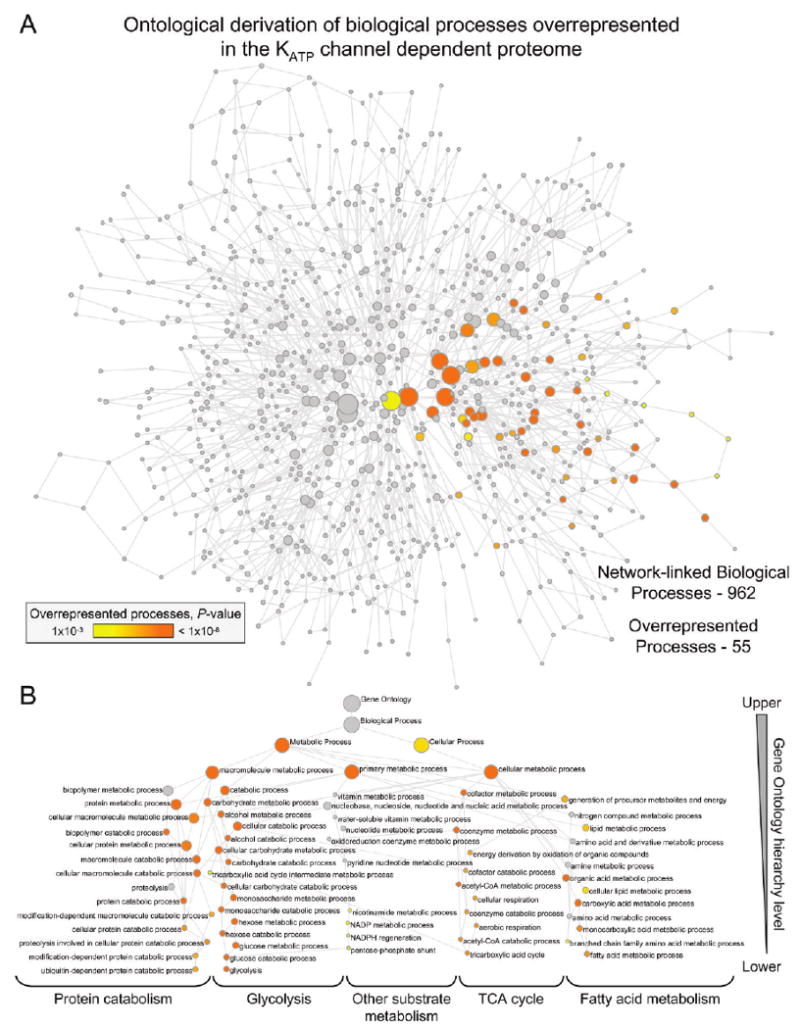
Biological processes linked to the KATP channel-dependent protein interaction network. (A) KATP channel-dependent protein interaction network was interrogated with the Cytoscape module BiNGO to identify network-linked Gene Ontology (GO) biological processes, including evidence of overrepresentation. The derived GO network consisted of 962 processes (nodes), of which 55 were significantly overrepresented (colored nodes, P < 0.001), with nodes connected by GO hierarchical relationships. Node size is proportional to the number of network proteins annotated to that biological process, while node shading represents a 5-log gradient of significant, false discovery rate-corrected P-values, with gray nodes not significantly overrepresented. (B) Hierarchical layout of overrepresented biological processes from panel A, with node size/color retained and identities indicated. Of note, all 55 colored nodes involve metabolic processes. Functional interdependencies within the GO hierarchy translated into significant overrepresentation by entire hierarchical branches, with the most significant nodes furthest down the hierarchy offering the strongest interpretive explanation. Processes linked to “Glycolysis”, the “TCA cycle”, “Fatty acid metabolism” and “Protein catabolism” demonstrated the greatest overrepresentation.
