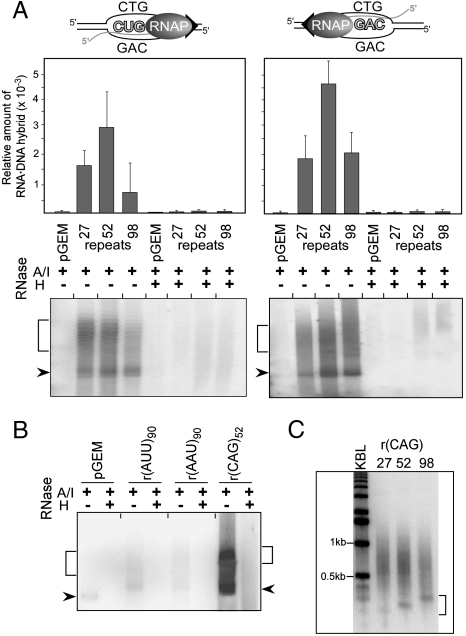
Fig. 1.
Stable RNA·DNA hybrids induced by in vitro transcription of CG-rich trinucleotide repeats. (A) RNA·DNA hybrids in transcribed CTG·CAG repeat tracts. Transcription reactions were treated with RNase A/I alone or in combination with RNase H prior to agarose electrophoresis. Plasmids containing 0, 27, 52, or 98 CTG·CAG repeats were all derivatives of pGEM3Zf(+). Arrowheads indicate migration of the supercoiled DNA; brackets indicate the position of relaxed plasmids. Plasmid-associated RNAs were quantified by scintillation counting and the efficiency of hybrid formation is expressed relative to the total amount of RNA prior to the RNase A/I/H treatment. (B) Lack of RNA·DNA hybrids in transcribed AT-rich (AAT·ATT)90 repeats. (C) Length of RNA·DNA hybrids formed at transcribed CTG·CAG repeat tracts of different lengths. Plasmid-associated RNAs were extracted from agarose gels, incubated with DNaseI and analyzed on a 1.2% agarose gel in denaturing conditions; KBL designates a 1 kb DNA ladder. The lengths of the shortest RNAs involved in RNA·DNA interactions, indicated by the bracket, correlate with the length of the CTG·CAG tracts.