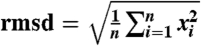Table 3.
Structural parameters of [(mepFlpGly)7]3 and other crystalline triple helices
| Parameter | [(mepFlpGly)7]3* | [(ProHypGly)10]3† | [(ProProGly)10]3‡ |
| resolution (Å) | 1.21 | 1.26 | 1.3 |
| interstrand hydrogen bond (Å) | 2.97 ± 0.06 | 2.88 | 3.04 ± 0.01 |
| helical pitch | 7/2 | 7/2 | 7/2 |
| ϕ, Xaa position (deg) | -76.0 ± 4.1 | -71.3 ± 1.4 | -74.5 ± 2.9 |
| ψ, Xaa position (deg) | 164.7 ± 3.9 | 161.5 ± 1.1 | 164.3 ± 4.1 |
| ω, Xaa position (deg) | 180.12 ± 2.7 | 172.3 ± 1.0 | 176.0 ± 2.5 |
| ϕ, Yaa position (deg) | -60. ± 1.5 | -56.9 ± 1.3 | -60.1 ± 3.6 |
| ψ, Yaa position (deg) | 151.3 ± 3.6 | 150.0 ± 1.1 | 152.4 ± 2.6 |
| ω, Yaa position (deg) | 176.0 ± 1.3 | 174.7 ± 1.1 | 175.4 ± 3.4 |
| ϕ, Gly (deg) | -71.5 ± 2.6 | -71.3 ± 1.6 | -71.7 ± 3.7 |
| ψ, Gly (deg) | 176.8 ± 4.3 | 174.2 ± 1.2 | 175.9 ± 3.1 |
| ω, Gly (deg) | 175.3 ± 2.3 | 178.8 ± 1.1 | 179.7 ± 2.0 |
| rmsd (Å)§ | — | 0.2195 | 0.2320 |