Fig. 1.
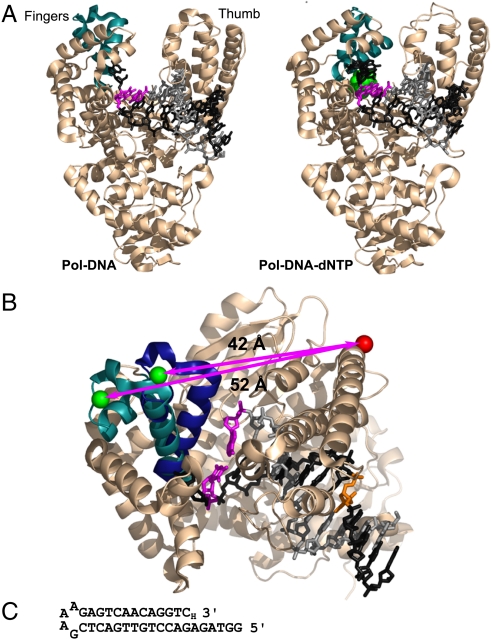
The fingers-closing conformational change in Pol I(KF). (A) The open Pol-DNA binary complex (PDB file 1L3U) and closed Pol-DNA-dNTP ternary complex (PDB file 1LV5) are illustrated using structural data from B. stearothermophilus (Bst) DNA polymerase (3), a close homologue of Pol I(KF). The α-carbon backbone of the protein is shown in Beige, except for the mobile segment of the fingers subdomain in Teal. The DNA template strand is in Dark Gray, the primer strand in Light Gray. The terminal base pair at the active site is Magenta, and the incoming complementary dNTP is Green. (B) Superposition of the open and closed structures shown in (A), viewed from above the polymerase active site. The mobile portion of the fingers subdomain (residues 680 to 714, equivalent to Pol I residues 732 to 766) is shown in Teal in the binary complex and Dark Blue in the ternary complex. The backbone structure of the rest of the protein, shown in Beige, is taken from the 1L3U PDB file but is essentially the same in both structures. The β carbons of the two side chains used as fluorophore attachment sites are shown in space-filling representation; residue 744 in Green, and residue 550 in red (Pol I residue numbers); the arrows indicate the distance between the Cβ positions in the open and closed conformations. The DNA primer-template is colored as in (A), except for the T(-8) position (Orange), which served as the attachment site for a dabcyl quencher in some experiments (see Fig. S1). The illustrations in (A) and (B) were made using PyMOL (DeLano Scientific). (C) DNA-hairpin oligonucleotide used in single-molecule FRET experiments. A dideoxy nucleotide (3′-H) prevents covalent addition to the 3′ end.