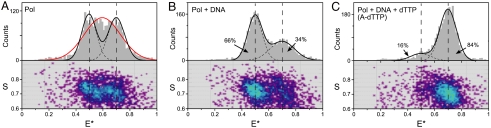
Fig. 2.
FRET-related histograms for Pol I(KF) complexes. (A–C) FRET-related histograms for (A) unliganded R550G744 Pol I(KF), (B) the Pol-DNA binary complex, and (C) the Pol-DNA-dNTP ternary complex (with a correct dNTP, forming an A-dTTP pair). The numbers of bursts in A, B, and C are comparable (2731, 2908, and 2598 events, respectively). The binary complex was formed by adding 100 nM DNA to Pol I(KF); the ternary complex was formed by adding 100 nM DNA and 10 μM dTTP to Pol I(KF). Each sample is described by two histograms: The lower shows a two-dimensional histogram of probe stoichiometry, S, vs. apparent FRET efficiency, E∗, for each diffusing molecule that contains both G and R fluorophores (i.e., molecules with 0.6 < S < 0.9; see SI Materials and Methods and refs. 14, 31). The upper panel shows a one-dimensional E∗ histogram (80 bins) of the molecules in the lower panel. In (B) and (C), the E∗ distributions were fitted to double-Gaussian distributions (black solid lines, sum of Gaussians; dashed lines, individual Gaussians) using an iterative process (see SI Materials and Methods). The percentage of the population in each Gaussian (after correction for the contaminating G550R744 protein) is indicated. The two dashed vertical lines mark the mean E∗ values of the main subpopulations in the binary (open) and ternary (closed) complexes. For the unliganded Pol I(KF) in (A), the red line shows the best fit by a single Gaussian; the black dashed lines show a fit to a constrained double-Gaussian distribution with the means and standard-deviations of the Gaussian peaks set to equal those of the open (〈E∗〉 = 0.5, σSN = 0.054) and closed (〈E∗〉 = 0.7, σSN = 0.060) complexes determined from the data in (B) and (C); neither of these strategies provide a good fit to the experimental data (see text). The residuals of all fits are in Fig. S5.