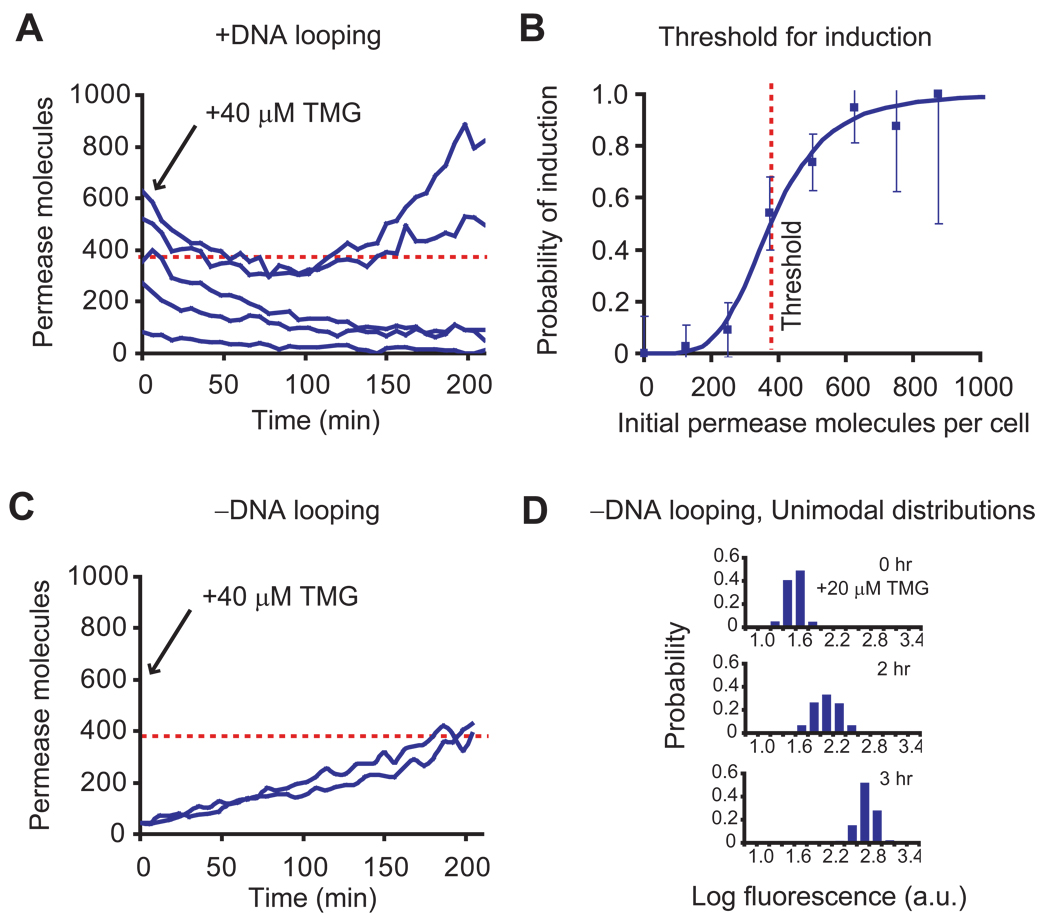
Figure 2.
Measurement of the threshold of permease molecules for induction. (A) Single cell time traces of fluorescence intensity, normalized by cell size, starting from different initial permease numbers. The initial LacY-YFP numbers are prepared through dilution by cell division of fully induced cells after removal of inducer. Upon adding 40 µM TMG at time zero, those cells with low initial permease numbers lose fluorescence with time as a result of dilution by cell division and photobleaching, while those cells with high initial permease numbers exhibit an increase in fluorescence as a result of reinduction. Permease molecule numbers are estimated from cell fluorescence (28). (B) The probability of induction of a cell within three hours as a function of initial permease number was determined using traces from 90 cells. The probability of induction, p, is fit with a Hill equation p = y4.5 / (y4.5 + 3754.5) for initial permease number, y. The threshold of permease numbers for induction is thus determined to be 375 molecules. Error bars are the inverse square root of sample size at each point. (C) To prove that complete dissociation of tetrameric repressor from two operators triggers induction, we constructed strain SX702 with auxiliary operators removed (no DNA looping). The figure shows single-cell traces of permease numbers in single cells grown in 40 µM TMG as a function of time. Unlike the looping strain SX700, the rapid induction of SX702 is no longer dependent on the initial number of permease molecules. This proves that phenotype switching is the result of a complete dissociation of the tetrameric repressor as shown in B. (D) In the absence of DNA looping, the entire population of strain SX702 rapidly induces in a coordinated manner from far below the threshold for a concentration as low as 20 µM TMG. DNA looping is necessary for bistability of the lac operon under these conditions.