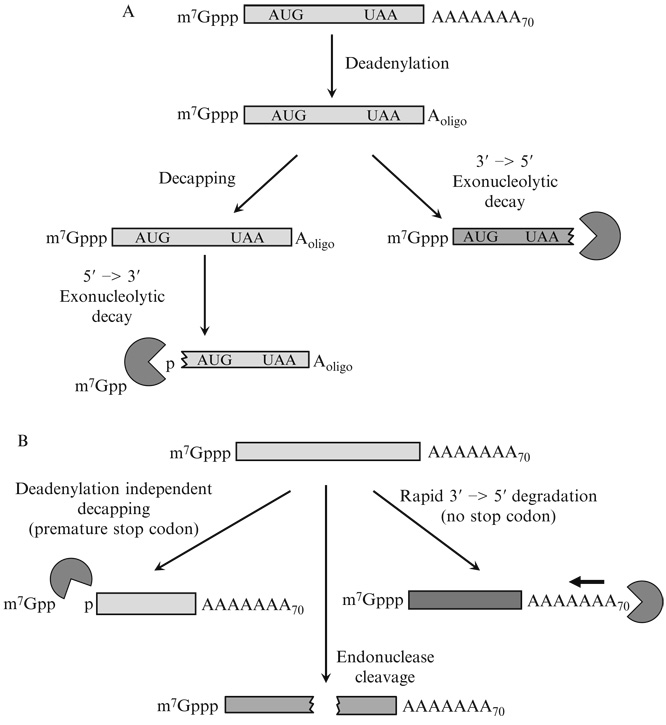
Figure 20.1.
Pathways of mRNA decay in yeast. (A) Model showing the two major routes for general mRNA decay in yeast. Normally, mRNAs are initially subjected to deadenylation followed either by decapping and 5′ to 3′-decay by Xrn1p or by 3′ to 5′-degradation by the exosome. (B) Three pathways known in yeast for bypassing the deadenylation requirement for mRNA turnover include nonsense-mediated mRNA decay (left), which causes deadenylation independent decapping, no-go decay (middle), which leads to endonucleolytic cleavage, and nonstop decay (right), which leads to rapid and continuous 3′ to 5′-degradation.