Figure 7.
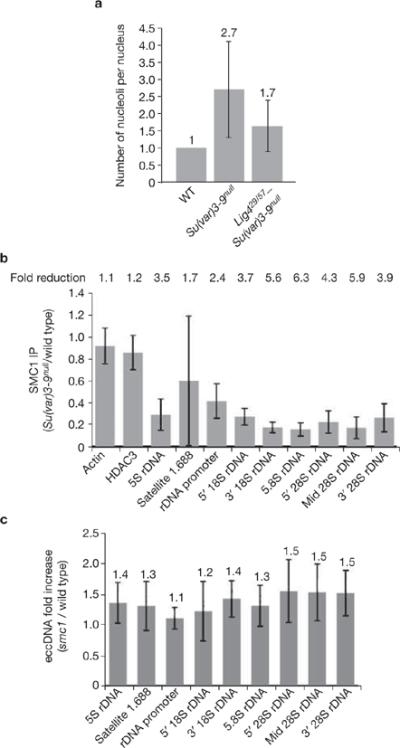
Effects of Ligase 4 and cohesin mutants on ectopic nucleolus and eccDNA formation. (a) Ligase 4 mutations partially suppress ectopic nucleolus formation in Su(var)3–9 mutants. Average nucleolus number of Lig4null–Su(var)3–9null polytene nuclei is 1.7 (n = 83), which is significantly lower (P <0.001) than the average 2.7 (n = 54) nucleoli observed in Su(var)3–9null mutant nuclei. Absolute values are indicated above each bar. (b) ChIP analysis shows reduced SMC1 levels at repeated DNAs in Su(var)3–9null chromatin, relative to wild type; fold reductions are shown (P <0.05 for all repeated DNA except 5S rDNA). Values are averages of four PCR reactions from two ChIP experiments. (c) The amount of eccDNA from satellite 1.688 and rDNA in smc1exc46l mutant tissues do not differ significantly from wild type. Absolute values are indicated above each bar. Error bars indicate s.d.
