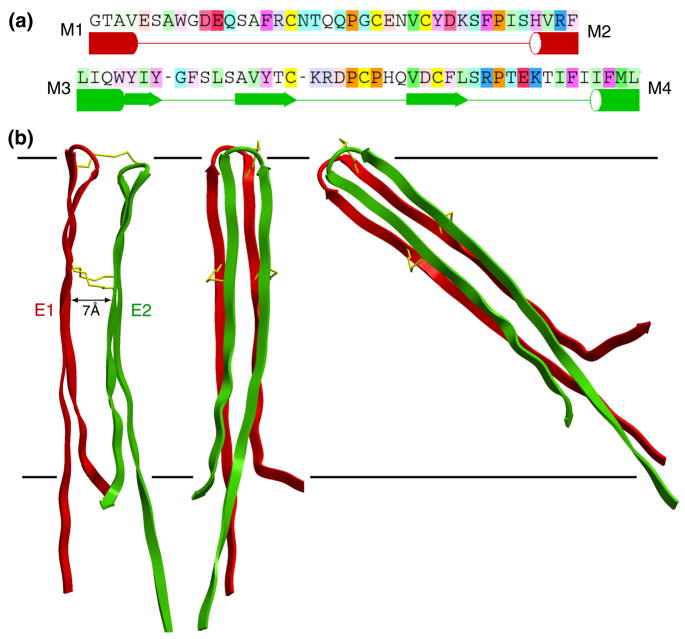
Figure 3.
(a) Sequences of loops E1 (top) and E2 and a statistical secondary-structure prediction with ICM software (in preparation). Tubes and arrows are predicted to be TM helices and β-sheet secondary structure, respectively. Note that the prediction for E2 shows partial β-strand structure. (The residue coloring comes from the alignment, and depends on both the conservation and the residue type as in Figure 4.) (b) Loops E1 (red) and E2 (green) belonging to a single connexin subunit. The left side shows a side view, while the middle and right sides are pore views. The separation between the loops is ~7 Å. The right side shows that the 35 amino acids of the complete loops would have to be tilted by ~50° to be accommodated within the 40 Å extracellular gap (represented by horizontal lines), if one assumes that they form extended β strands. The cysteines forming disulfide bonds are indicated by gold sticks.