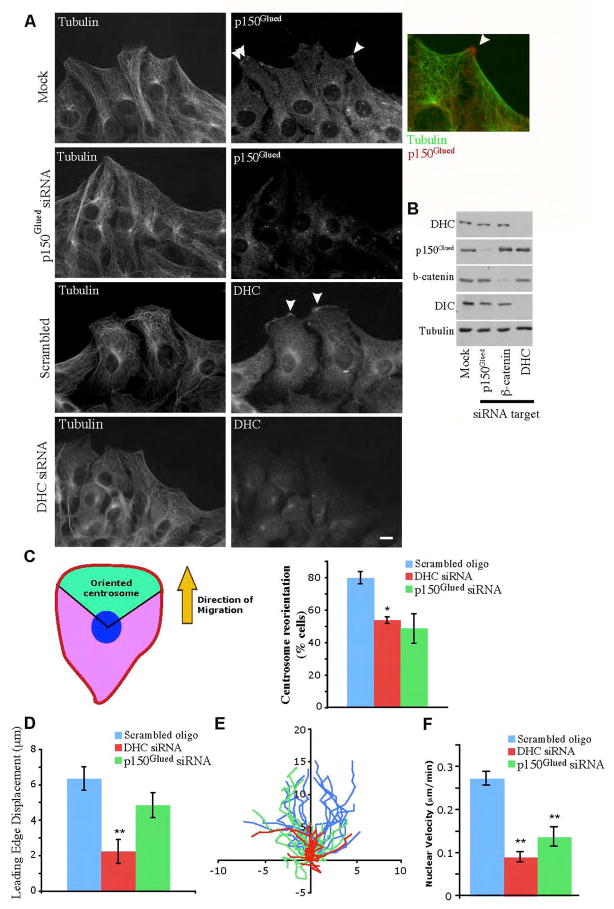
Figure 1.
Dynein and dynactin localize to the leading edge of migrating cells and promote centrosome reorientation, protrusion, and nuclear migration. (A) Dynein and dynactin concentrate in cortical patches at the leading edge of migrating cells. NIH/3T3 cells that were mock-transfected or transfected with siRNAs targeting p150Glued or DHC were grown to confluency and the monolayer was wounded. Six hours after wounding, cells were fixed and stained for α-tubulin and either p150Glued (top) or DIC (bottom). Mock-transfected cells show patches of dynein and dynactin at the leading edge of migrating cells (arrowheads). Inset, overlay of cortical patch of p150Glued (red) and projecting MTs (green). Bar, 10 μm. (B) Western blot of protein lysates from cells that were mock-transfected, transfected with siRNAs targeting p150Glued, DHC, or β-catenin, a protein that binds to dynein (Ligon et al., 2001) consistently show knockdown of target protein levels by 75–95%. Levels of DIC decrease in DHC knockdown cells, suggesting the dynein complex is being destabilized. In contrast, knockdown of β-catenin, which binds to DIC but is not part of the dynein motor complex, does not destabilize DIC. (C) Centrosome reorientation is inhibited in dynein and dynactin knockdown cells. Orientation of centrosomes 6 hours after wounding. Centrosome orientation was determined by measuring the position of centrosomes, as determined by γ-tubulin staining, in relation to Hoescht stained-nuclei. Centrosomes in the forward facing 120 degree sector (green zone) were scored as reoriented. (D) Leading edge extension is inhibited in dynein knockdown cells. Displacement of the leading edge over 45 min of wound healing was measured (n=15). (E) Tracks of nuclei centroids during migration of fibroblasts over 45 min. Paths are oriented so cells are migrating towards the top of the graph, each path starts at (0,0). Axis labels are in microns. (F) Rates of nuclear movement is decreased in dynein and dynactin knockdown cells. Nuclear velocity during migration was decreased in DHC and p150Glued siRNA cells, compared to mock transfected cells (n=15). Error bars indicate SEM; * indicates p<0.05.; ** indicates p<0.005.