Abstract
Plants respond to environmental stresses by altering transcription of genes involved in the response. The chromatin modifier ATX1 regulates expression of a large number of genes; consequently, factors that affect ATX1 activity would also influence expression from ATX1-regulated genes. Here, we demonstrate that dehydration is such a factor implicating ATX1 in the plant's response to drought. In addition, we report that a hitherto unknown Arabidopsis gene, At3g10550, encodes a phosphoinositide 3′-phosphatase related to the animal myotubularins (atMTM1). Myotubularin activities in plants have not been described and herein, we identify an overlapping set of genes co-regulated by ATX1 and AtMTM under drought conditions. We propose that these shared genes represent the ultimate targets of partially overlapping branches of the pathways of the nuclear ATX1 and the cytoplasmic AtMTM1. Our analyses offer first genome-wide insights into the relationship of an epigenetic factor and a lipid phosphatase from the other end of a shared drought responding pathway in Arabidopsis.
Key words: response to drought stress, histone modification, ATX1, plant phosphoinositide 3-phosphatase, AtMTM, lipid signaling and chromatin modification
Introduction
Chromatin structure underlies genome activity by limiting the accessibility of the transcriptional machinery to the gene. Interactions of DNA with the core histones determine the first level of structural organization represented by the 11 nm chromatin fibers. Chemical modifications of the histone amino acid residues may influence this structure. Factors that modify chromatin and, as a result, influence gene activity are defined as epigenetic.1 The paradigm for epigenetic mechanisms is the function of the counteracting Polycomb Group/Trithorax (PcG/TrxG) complexes.2,3 PcG and TrxG activities establish specific modifications at the histone-tails providing ‘tags’ that are ‘read’ by factors recognizing modified residues. These tags may serve as epigenetic marks extending the informational potential of the genetic code.1–4 Initially, PcG/TrxG-driven epigenetic mechanisms were associated exclusively with animal developmental processes. Currently, PcG/TrxG factors are thought to underlie genome plasticity implicated in adaptation processes, in centromeric- and telomeric-silencing, in cell cycle regulation, in senescence, disease and cancer.5
PcG/TrxG related factors controlling developmental processes in plants are studied by many plant research laboratories6 but evidence for the roles of PcG/TrxG and the epigenome in environmental adaptation and responses to biotic and abiotic stresses is only now beginning to emerge.7,8 For example, the Arabidopsis Trithorax homolog, ATX, participates in the salicylic acid/jasmonic acid response pathways7 and regulates transcription from over 130 biotic and abiotic stress response genes including members of the TIR-NBS-LRR families of disease resistance proteins, mildew-resistant factors, lectins, major latex proteins, LEA and Bet v I allergen family proteins and over 20 genes encoding chaperones and heat shock proteins.9 A recent study reported changes in the histone H3-lysine modifications under drought.10
The ability of ATX1 to coordinate transcript levels of a large number of diverse genes suggests that it acts as a ‘master-regulator’ By modifying targets positioned at critical nodes of branching gene networks, chromatin factors can provide flexibility and rapid-responses to challenges by selectively modulating transcriptional intensities of only a few primary targets.7,11–13
Biochemically, ATX1 is involved in tri-methylating lysine 4 of histone H3 (H3K4me3),14,15 a modification associated with transcriptionaly active genes. In addition, ATX1 can bind specifically the lipid phosphatidyl-inositol 5-phosphate (PtdIns5P) and the two co-regulate a shared set of genes.9,16 Based on these data we proposed a model according to which elevated level of cellular PtdIns5P negatively influence the activity of ATX1.9 Because PtdIns5P concentration increases in response to hyperosmotic stress in yeast,17 human,18,19 Chlamidomonas and carrot culture cells,20 it was logical to suggest that the stress-elevated PtdIns5P level could affect ATX1 activity linking, thus, epigenetic regulation with lipid signaling. Proving this model, however, is challenging because PtdIns5P is a minor component of the cellular phospholipid pool only recently discovered in osmotically stressed cells.17–20 Difficulties in measuring cellular PtdIns5P and overlapping PtdIns5P and PtdIns4P peaks in HPLC profiles have been the main obstacles to identifying PtdIns5P in Arabidopsis cells21 and to testing the proposed model experimentally.
However, an alternative approach to manipulate the cellular PtdIns5P, routinely used in animal cells, is to alter the enzyme activity producing PtdIns5P. Myotubularins (MTMs), belong to the large family of dual serine-threonine phosphatases that can generate PtdIns5P from PtdIns3,5P2 by specific dephosphorylation at position 3 of the inositol ring.22–24 Ectopic expression, or depletion, of a myotubularin gene is used as a tool to modulate the concentrations of endogenous substrates (PtdIns3P and PtdIns3,5P2) and respective products (PtdIns and PtdIns5P). Disrupted cellular homeostasis of these lipids is linked with severe neuron-muscle degenerative diseases.24–28
Myotubularin activities of plant origin, however, have not been reported. Hereby, our first goal was to identify a putative counterpart of a human myotubularin gene in Arabidopsis (AtMTM) and to determine whether it has a 3′-phosphatase activity with PtdIns3,5P2. Transgenic lines stably overexpressing a myotubularin homologous gene (OX-AtMTM) were constructed and analyzed for possible effects upon transcript accumulation in a genome-wide context. Changes in transcript levels were measured under irrigated (control) and drought (stress) conditions and compared with the profiles displayed in the atx1 mutant background. We reasoned that a set of common drought-responsive genes identified from the atx1 and the OX-AtMTM data sets would represent genes co-regulated by the chromatin modifier and the lipid phosphatase outlining, thus, the overlapping branches of pathways involving both ATX1 and AtMTM.
Results
Genome-wide transcript accumulation in watered and drought stressed plants in the three different genetic backgrounds.
Microarrays were used for genome-wide analysis of wild type (Col), atx1 and AtMTM overexpressing (OX-AtMTM) plants under non-stress (irrigated) and upon stress (water withdrawal) conditions. Wild type non-stressed and stressed samples had correlation coefficients of 0.95 and 0.97 respectively; the nonstressed and stressed atx1 had correlation coefficients of 0.93 and 0.92, respectively; and the OX-AtMTM untreated and stressed plants had correlation coefficients of 0.86 and 0.95, respectively (see scatter plots in SF1). Consistently, our analyses detected about 65% (∼15,600) of all Arabidopsis genes expressed at this particular developmental stage.
The data mining methods and the criteria used to define genes with robust change in transcript levels are described (Methods). To verify microarray data, real time PCR assays were performed on several genes randomly selected from the different microarray samples (Fig. 1).
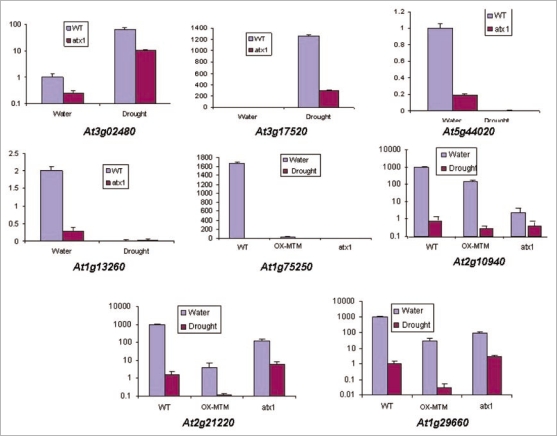
Figure 1.
Real-time quantitative RT-PCR analysis of drought-responding genes. Samples were randomly selected to represent genes with altered transcript levels in the different experimental samples and in the wild type controls. Y-axis shows the relative transcript levels levels compared with Ubiquitin. The At3g02480 gene encodes an ABA-responsive protein and the At3g17520 gene encodes a LEA protein; At5g44020 encodes a class B acid phosphatase; At1g13260 and At2g21220 encode proteins implicated in the brasinosteroid and auxin responses, respectively; At1g75250 is a transcription factor from the Myb-family, while At2g10940 and At1g29660 encode lipid transfer and lipid hydrolyze activities, respectively.
Among the approximately 15,600 detected genes, 3,856 genes responded to drought stress by altering transcript levels more than three-fold. Cutoff for the log2 signal ratio is 1.5 (∼three-fold change) (p < 0.05): 2,345 genes decreased and 1,511 increased transcript levels upon drought treatment.
All data from the microarray profiling are deposited in the NCBI Gene Transcript levels Omnibus series GSE15577.
Misexpressed genes in the atx1 background overlap with genes changing transcript levels in the wild type upon stress.
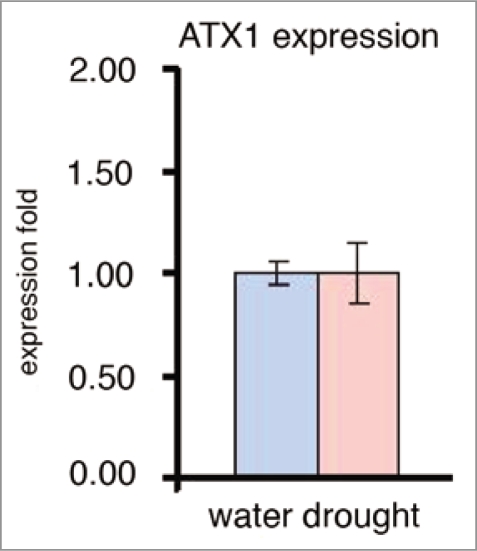
As a first step in our analyses, we examined whether the transcript levels of the ATX1 gene is affected by the drought stress. The result showing that ATX1 transcript levels is not influenced by drought conditions (Fig. 2) is important because it suggests that altered transcript levels of ATX1-regulated genes in wild type stressed plants would be due to changed ATX1 activity, not to a change in ATX1 transcript levels.
Figure 2.
Transcript levels of the ATX1 gene upon exposure to water withdrawal. Real-time RT-PCR assay of the ATX1 gene transcript levels in non-stressed wild type leaves and in leaves exposed to air for two hours (see text for details).
Wild type and atx1 mutant plants were subjected to water withdrawal stress (see Methods) and the data from the genome-wide gene were analyzed by several different comparisons. After comparing the stressed atx1 dataset with the stressed wild type sample we identified 2,810 differentially transcribed genes (Suppl. Table 1). It is important to note, however, that this large gene set reflects the compounded effects of two variables: ATX1-loss of function and the stress. Thereby, to distinguish a possible effect of the dehydration upon the activity of ATX1, we performed two additional comparisons: first, the genes with altered transcript levels when the atx1 watered and atx1 dried sets were analyzed among the 920 genes, 630 increased, 290 decreased transcription (Suppl. Table 2). These genes reflect the effects of dehydration in the atx1 background. We asked next whether there would be genes in non-stressed atx1 plants that would be shared with the drought-stressed wild type plants. Such an analysis could identify the gene set that in non-stressed ATX1-loss-of-function plants behave as wild type plants under stress reflecting, thus, the effects of the stress upon the activity of ATX1.
Cluster (overlap) analysis identified 686 common genes (Suppl. Table 3). The significance of the overlap was computed via chisquare (p = 0). In the overlapping set, 446 genes were upregulated, 240 genes were downregulated. Thereby, these overlapping gene subsets represent genes that alter their transcript levels in the wild type upon stress similarly to the same genes in non-stressed atx1 mutant plants. Statistical analysis indicated that the magnitudes of the changes were also correlated: the Pearson correlation coefficient is 0.78 (p < 0.0001, n = 446) for up-and 0.65 (p < 0.0001, n = 240) for downregulated genes. Real-time quantitative RT-PCR analyses of a sample of these genes confirmed microarray data (Fig. 1). Because the genes that changed transcript levels under drought stress in the wild type similarly changed transcription in the atx1 background and because ATX1 transcripts do not decrease upon dehydration, it is logical to conclude that drought stress negatively impacts (inhibits) the activity of ATX1.
Characterization of the ATX1-regulated/drought-induced shared functions.
The 686 overlapping genes were examined individually for their subcellular localization and function as determined in the NCBI and TAIR databases (Table 1). Remarkably, 204 genes (about 30%) in the ATX1-regulated drought-responding fraction encoded plasma membrane and cell wall associated functions. These represented ABC-type and metal transporters, potassium-channel and transmembrane proteins, cytochrome P450 family members and enzymes involved in membrane transfers of sugar and lipids. Genes encoding protein kinases, phosphatases and putative components of signal transduction pathways were also represented in the plasma membrane/wall associated shared fraction. Forty-nine genes represented chloroplast-associated genes, while only four were associated with the mitochondria. Ten percent of the shared fraction encoded nuclear functions including 52 transcription factors: 11 members of the MYB, 10 Zinc-finger, eight of the NAM, four of the WRKY, and three of the CCAAT-box and the bHLH families (Suppl. Table 3). It is noted that these genes represent a rather limited subgroup of the Arabidopsis transcription factor families and that members of some of the largest families, like the MADS-box, or the AP2-box, families were absent in the overlap.
Table 1.
Association of the shared ATX1-regulated/drought-induced/OX-AtMTM affected genes with cellular structures and response mechanisms
| Genes with altered transcript levels shared by the atx1 and wt-stressed samples | |||||||
| Total genes in the overlap | Endomembrane/wall genes | Chloroplast genes | Nucleus genes | Response mechanisms | Cytoplasm and metabolism | Unspecifieda genes | Unknownb genes |
| 687 (100%) | 204 (30%) p ≤ 0.000006 | 49 (6%) | 65 (10%) | 59 (9%) | 86 (13%) | 147 (22%) | 77 (10%) |
| Shared genes in the three backgrounds (atx1/OX-AtMTM/wt-stressed) | |||||||
| Total genes in the overlap | Endomembrane/wall genes | Chloroplast genes | Nucleus genes | Response mechanisms | Cytoplasm and metabolism | Unspecifieda genes | Unknownb genes |
| 140 (100%) | 63 (45%) p ≤ 0.005 | 6 (4%) | 12 (9%) | 33 (23%) | 20% | 13% | 10% |
Proteins with no assigned subcellular structure.
Expressed proteins with unknown predicted functions and no assigned subcellular structure. These entries are a subfraction of the group Unspecified. GO background and significance analysis via AmiGO and GOEAST, least significant p value of both tools was taken. The sum of indicated genes is over 140 because some genes were identified by more than one feature; for example, response genes may be associated with membranes, with the nucleus or be in the cytoplasm.
Eighty-six known and predicted genes were involved in metabolic and cytoplasmic functions, while ∼57 genes were involved in biotic and abiotic stress-responses. It is interesting to note that activities associated with auxin, gibberellin, and brasinosteroid responses were present only in the downregulated fraction, while 45 genes encoding activities responding to ABA, cold, salinity and drought were found only in the upregulated set; five HEAT SHOCK (HS) and eight LATE EMBRYOGENESIS ABUNDANT (LEA) genes were also present only in the upregulated group. About 35% of the shared genes did not have predicted association with cellular substructures and/or encoded expressed proteins with unknown function and unknown compartmentalization (Table 1; Suppl. Table 3).
Myotubularin-like genes in arabidopsis.
A missing factor critical for supporting a possible role of PtdIns5P in drought responses was the lack of evidence for enzyme activity capable of producing the lipid in Arabidopsis. In animals, the MTM phosphatases generate cellular PtdIns5P.24 To establish whether myotubularin-like genes exist in Arabidopsis, the genome was searched for genes with similarity to the amino acid sequence of the human MTMR2. Two putative myotubularin (AtMTM) homologs encoded by the At3g10550 and At5g04540 genes (referred here as AtMTM1 and AtMTM2, respectively) were identified. They are highly related between themselves (77% identical, 85% similar, e = 0.0) and to the human MTMR2 (38% identical, 59% similar, 4e−70) containing similar domains (Fig. 3A). All the consensus amino acids critical for the enzyme activity of the human myotubularin33,34 are conserved in the two plant homologs. The results reported below are for AtMTM1.
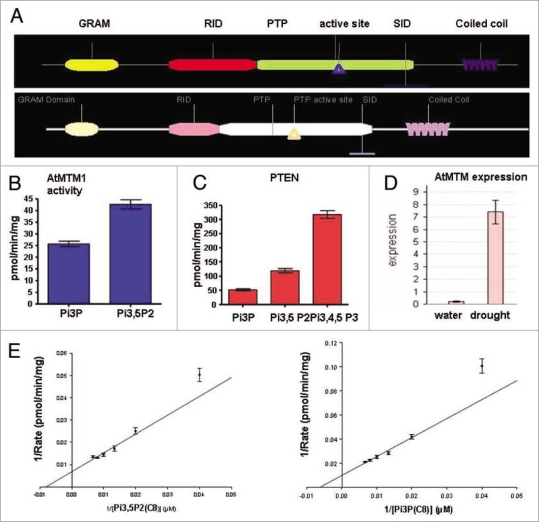
Figure 3.
The Arabidopsis Myotubularin homolog At3g10550: domain structure, enzyme activity and AtMTM1 transcript levels under irrigated and water deficit conditions. (A) Structure of the Arabidopsis At3g10550 protein AtMTM (top) and the human MTMR2 (bottom). Domains are as defined in Begley and Dixon;33 (B) Phosphoinositide 3′-phosphatase activity of the recombinant protein containing RID-PTP-SID domains with PtdIns3P and PtdIns3,5P2 as substrates; (C) activity of PTeN (phosphatase and tensin) assayed in parallel as a positive control; (D) Real-time RT-PcR assay of the AtMTM1 gene transcript levels in non-stressed wild type leaves and in leaves stressed by exposure to air for two hours; (E) Lineweaver-Burk curves for Ptdins3P and Ptdins3,5P2 used as substrates for atMTM: Km = 146 µM and Vmax = 0.12 nmol/min/mg for PI(3)P; Km and Vmax for PI(3,5)P2 (C8): 101 µM and 0.16 nmol/min/mg.
To test whether an Arabidopsis myotubularin-like protein is capable of dephosphorylating PtdIns3,5P2 to produce PtdIns5P, we constructed recombinant GST-tagged fusion clones for transcript levels in bacterial cells. Attempts to produce the entire protein resulted in very low yields insufficient for biochemical studies. However, myotubularins containing the catalytic site and the flanking RID and SID domains display phosphatase activity;22–24 thereby, we constructed a GST-tagged clone encoding the catalytic (PTP) domain plus the flanking (RID and SID) domains of the At3g10550 gene. The recombinant protein was affinity purified and tested for phosphoinositide 3′-phosphatase activity. Following published procedures28,31 we established phosphatase activity for AtMTM with both PtdIns3,5P2 and PtdIns3P as substrates (Fig. 3B). Like the mammalian myotubularins, the plant protein displayed 3′-phosphatase activity against both substrates and, like the human MTMR2, its activity was higher with PtdIns3,5P2 than with PtdIns3P22,28 indicating that the bi-phosphate is a preferred substrate for AtMTM1 as well. PTEN activity was assayed in parallel as a positive control (Fig. 3C and D).
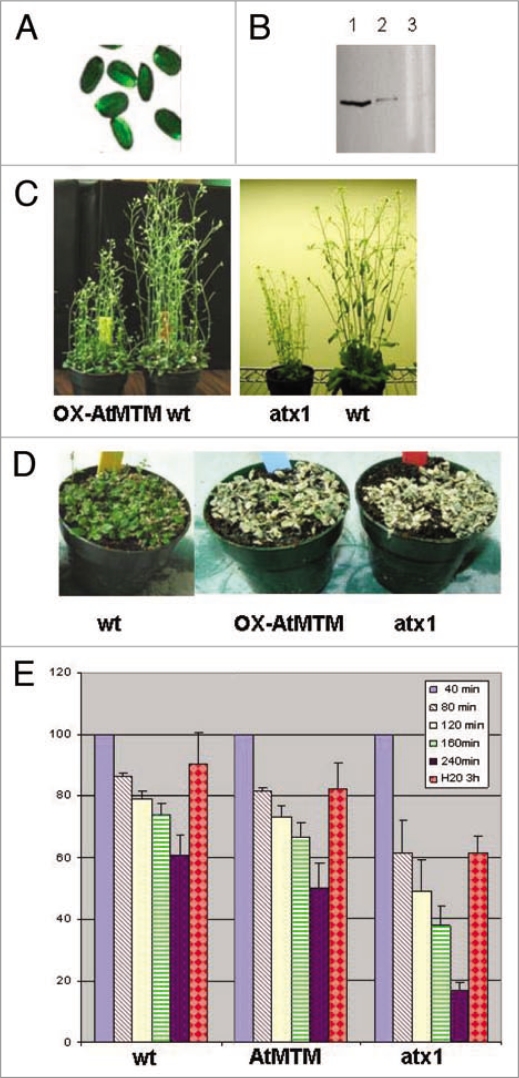
Unlike ATX1, the transcript levels of the AtMTM1 gene increased significantly upon water withdrawal (Fig. 3E). To analyze its possible involvement in the plant's response to dehydration, we cloned the entire At3g10550 coding sequence under the 35S promoter and generated transgenic Arabidopsis lines overexpressing AtMTM1 (OX-MTM1) (Fig. 4A and B). During development, OX-AtMTM plants did not show detectable differences from wild type, except that at the flowering stage they remained shorter; atx1 plants also remained shorter than wild type at this developmental stage (Fig. 4C).
Figure 4.
OX-AtMTM lines and responses of OX-AtMTM and of atx1 mutant plants to drought. (A) Transcript levels of GFP on seed coats, reflecting the overexpression of AtMTM1. Overexpressing plants can be easily identified under fluorescent microscope; (B) western blots of two stably HA-AtMTM1 overexpressing lines showing different levels of ectopic AtMTM1 accumulation (lanes 1 and 2) as detected with HA antibody. Lane 3 is extract from wild type plants as a negative control; (C) Stably transformed AtMTM1-overexpressing and wild type plants grown under regular irrigation (left-hand). atx1 and wild type plants grown under exactly the same conditions (right-hand); (D) wild type, OX-AtMTM, and atx1 plants not watered for five days followed by two days of irrigation; (E) Water loss in detached leaves as a function of time exposed to air. Changes in tissue mass upon air exposure for the indicated periods of time, followed by submersion in water for three hours are shown as percentage of the weight of starting tissue material taken as 100% for each sample. Bars are averages of three independent measurements. Changes were statistically significant, t-test p > 0.01 for all samples.
Next, we examined the responses of the atx1 and OX-AtMTM mutant plants to water withdrawal and their recovery upon subsequent re-hydration in both potted plants and in detached leaves. In contrast to wild type, stressed plants from both mutant backgrounds failed to recover after being kept in soil without watering for five days followed by irrigation for three consecutive days (Fig. 4D). Similar responses were displayed by detached leaves from the two mutant samples. When exposed to air for the indicated periods of time, water loss was highest in leaves from atx1 mutant plants (more than 80% water content loss, s.d 2.7%). Wild type leaves lost about 40% (sd 6.6%) during the four-hour treatment, while OX-AtMTM lost about 50% (s.d 7.9%). Upon submersion in water for three hours, leaf weights were measured, and the data calculated as a percentage of the initial fresh weights. The recovery was highest in wild type leaves (90%), less in atx1 leaves (61%), and intermediary in OX-AtMTM leaves (82%) (Fig. 4E).
The most important result, relevant for our further studies, was that ATX1 and AtMTM1 were involved in the same, or largely overlapping, response pathway(s) as suggested by the apparently similar responses to the stress displayed by atx1 and OX-AtMTM plants. To test this hypothesis, we analyzed the transcript levels of genes in plants producing an excess of phosphatase and compared these with the genes from the atx1 samples implicated in the response.
Transcript profiling of the genes in the OX-AtMTM background.
Microarray profiling of non-stressed OX-AtMTM samples revealed that about 80 genes with altered transcript levels compared to the wild type (Suppl. Table 4). Comparison of the OX-AtMTM and atx1 data from plants grown under control watered conditions revealed only seven common genes misexpressed above the cutoff: At1g015290, encoding a MYB transcription factor, At1g17020, a putative oxidoreductase, At1g56600, a galactinol synthase, At3g14440, epoxycarotenoid dioxygenase, At3g12580, a heat shock 70 protein, At3g60140, a glycosyl hydrolase and At4g29780, an expressed protein of unknown function. This result is important because it indicates that in non-stressed plants, OX-AtMTM and atx1 function in largely non-overlapping pathways. However, the situation changes upon stress (see further below).
It is worth noting also that the relatively small number of misexpressed genes in the OX-AtMTM background is, most likely, due to the limiting substrate (PtdIns3,5P2) concentration in non-stressed cells. However, cellular PtdIns3,5P2 levels rise upon osmotic stress17,20,21 suggesting that effects from higher 3′-phosphatase levels could be better revealed in osmotically stressed plants. Subsequent microarray analysis of droughtstressed OX-AtMTM plants identified more than three thousand genes with altered transcript levels. However, as discussed above for atx1, these genes represent a compound fraction containing both stress-stimulated and AtMTM1-affected genes. To identify differentially expressed AtMTM1-dependent genes within the general fraction responding to the stress, we analyzed the overlap between the gene-sets of wild type stressed and OX-AtMTM-stressed plants. About 570 common genes showed a difference in transcript levels above the cutoff in OX-AtMTM stressed and wild type samples: 95 genes up, 478 genes downregulated in myotubularin overexpressing plants in response to drought (Suppl. Table 5).
Analysis of these genes according to their association with cellular substructures and involvement in biotic and abiotic response mechanisms revealed functional distributions similar to the atx1-drought responding fraction (see above). Many genes encoded membrane- or wall-associated proteins, including a variety of integral or peripheral membrane proteins (Suppl. Table 5). About 20 genes encoding cytoskeletal functions were misexpressed in the OX-AtMTM drought-induced sample in contrast to only two genes from this group found in the shared atx1-wild type drought response fraction (see further below).
Overlapping genes in the atx1 and in the stressed OX-AtMTM and wild type backgrounds represent a common target.
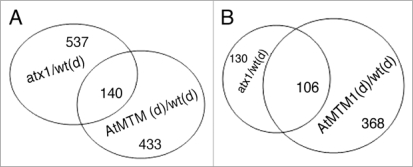
Overlap analysis identified about 140 genes shared among the three data sets (atx1-watered, OX-AtMTM-dry, and wild type-dry; chi-square p = 0, Suppl. Table 6, Fig. 5A). These genes represent a distinct stress-responding genome fraction targeted by a mechanism involving both ATX1 and AtMTM. Analysis of this common fraction revealed two important correlations: first, all genes up or downregulated in the atx1 sample were similarly up or down affected in the OX-AtMTM/wild type stressed samples. The patterns were confirmed by the real-time RT-PCR analyses of a few shared genes (Fig. 1). The results imply that overproduction of AtMTM protein resulted in gene transcript patterns similar to the patterns displayed by plants with disrupted ATX1 activity.
Figure 5.
Venn diagrams of the atx1/wt-stressed and the OX-AtMTM1/wt-stressed fractions. (A) Overlapping genes with significantly altered transcript levels in the atx1 and in stressed wild type backgrounds; (B) Overlap of downregulated genes in the atx1, OX-AtMTM and wild type drought-stressed samples.
Second, the downregulated genes in the overlapping set represented almost 80% of the genes repressed by dehydration stress in the atx1 and OX-AtMTM backgrounds (Fig. 5B; Suppl. Table 6). About 40% of the common targets represent genes encoding cell wall modifying and plasma membrane associated activities (Table 1). We note that transcripts from many genes involved in ethylene, auxin and gibberellin responses were similarly downregulated by drought treatment, by excess of AtMTM1, and by the ATX1-loss of function (in atx1), while genes involved in the responses to abscisic acid, cold and heat shock were found in the upregulated data sets.
Discussion
By modulating the structure of vast chromatin domains, chromatin modifiers play major roles in transcriptional reprogramming during plants' development and/or in response to environmental cues. ATX1 influences transcription from diverse classes of Arabidopsis genes including a subset of genes responding to water deficit stress. Due to the low enzyme activity of recombinantly expressed trithorax proteins in vitro,35 direct assays of the ATX1 biochemical activity are unfeasible. Here, as an alternative we use a genomics approach to analyze changes in ATX1 activity as reflected by changed transcript levels of ATX1-regulated/ genes. The overlap of the genes misexpressed in atx1 mutant plants (non-stressed) with wild type genes changing transcript levels upon water withdrawal defined a set of shared ATX1-regulated—stress-responding genes. It deserves to be noted also that ATX1-dependent genes may be still drought responsive in the absence of ATX1 (Fig. 1 and the array data) supporting further the idea that the effects of chromatin modifications upon gene transcript levels are secondary to the effects of transcription factors.7,14 Consistent with the idea that epigenetic regulators do not establish or silence transcription states, but only modulate the level of transcript levels, our results illustrate that under stress, in the absence of ATX1 or upon elevated phosphatase the transcript levels of the shared genes change in the same direction (up or down) without completely shutting transcription off.
Genome-wide transcript levels profiling and cluster (overlap) analyses are among the best available tools for outlining common pathways.9,16 Our data revealed that 686 shared genes changed transcript levels more than three-fold in atx1 mutant non-stressed plants and in wild type plants subjected to water withdrawal (the Pearson correlation coefficient is 0.95, p < 0.0001, n = 686) indicating that these shared genes reflected lowered ATX1 activity in the wild type upon stress. Another potentially important correlation displayed by the misexpressed gene subsets in the atx1 and the AtMTM1 backgrounds under normal (watered) conditions is that only seven genes were shared between the two activities. By contrast, ∼140 common genes were affected by water withdrawal, reflecting a mechanism involving both ATX1 and AtMTM. We suggest that AtMTM1 and ATX1 go ‘different ways’ under normal conditions. Upon drought stress, however, specific branches of their pathways overlap to target a shared set of genes illustrating, thus, a cross-talk between ATX1 and AtMTM1. Responding similarly to the ATX1-loss of function, to the excess of AtMTM1, and to the stress from water deprivation, these genes provide in vivo evidence for a biologically relevant pathway connecting all three factors. Furthermore, our data support the conclusion that higher MTM levels, as well as stress, negatively affect ATX1 activity.
Within the fraction shared by the three data sets, about 80% of the genes are downregulated, while 20% are upregulated (Fig. 5A). In the shared set, two groups of genes stand out (Table 1): first, about 40% of the shared targets encode cell wall- and membrane-associated functions indicating that these activities are preferentially targeted by an ATX1-AtMTM1-involving mechanism. It is tempting to suggest that this mechanism specifically modifies the plasma membrane and the cell wall during the cellular response to dehydration stress; second, about 30 shared genes involved in responses to other stimuli were downregulated. These included auxin, brasinosteroid and gibberellin response genes. By contrast, ∼22% in the shared upregulated fraction (Suppl. Table 6) represented genes encoding abscisic acid, cold, or dehydration-related functions. We note also that both AtMTM1 and ATX1 have roles beyond the common dehydration stress-signaling pathway. About 20 genes for cytoskeletal proteins were misexpressed in the OX-AtMTM-stressed sample but only two genes from this group (the calmodulin-binding, AT2G41090, and the tubulin gene, AT1G20010) were found in the atx1-wild type drought responding fraction. We conclude that upon stress, ATX1 and AtMTM1 participate in partially overlapping mechanisms provoking specific responses of drought responsive genes in Arabidopsis.
It is important to emphasize also that it is not possible to correlate directly the stress phenotypes resulting from the ATX1 loss-of-function or from the ectopic transcript levels of AtMTM1 with those resulting from the overexpressed, or knocked down, function of any particular drought-responding gene. For example, the gene for the dehydration stress response transcription factor DREB2A36,37 is upregulated in atx1 plants but the mutants are more susceptible to the stress. Clearly, the interactions among the various activities affected by the ATX1 (the numerous misregulated transcription factors, the metabolic, cell-structure and wall proteins, in particular)38 are complex making it impossible to link directly a phenotype to any particular gene effect. The relevant information, from the stress-induced phenotypes is that atx1 and OX-AtMTM mutants respond similarly to the stress consistent with the model that their pathways partially overlap under drought conditions.
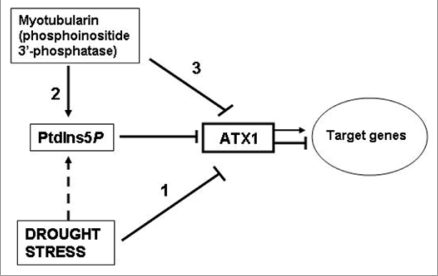
Earlier, we reported that ATX1 binds PtdIns5P via its PHD domain and that the two might participate in the same signaling pathway.9 Central to the model is the hypothesis that dehydration effects are mediated by PtdIns5P, whose levels rise under stress17–20 and that increased PtdIns5P negatively affects ATX1 activity (Fig. 6).
Figure 6.
A model involving AtMTM and ATX1 activities in the drought response. This model builds on and extends an earlier model suggesting that the lipid ligand PtdIns5P increases upon osmotic stress and negatively regulates ATX1 activity.9 Water withdrawal negatively affects ATX1 activity (bar 1); AtMTM1 has phosphatase activity generating the ligand (bar 2); excess cellular AtMTM1 affects transcript levels of the shared target genes like loss of ATX1 function does (bar 3). The model assumes that the effects of AtMTM1 and drought stress upon ATX1 activity are mediated by the lipid. however, the increase of PtdIns5P under stress in Arabidopsis remains to be shown (broken bar); Arrows and T-shaped bars indicate activation and repression events, respectively.
Whether cellular Ptdins5P levels rise under stress and in MTM1-overexpressing cells still remains to be proven. However, as shown earlier in animal cells,39 ectopic expression of enzymicaly active myotubularin is a tool to raise endogenous Ptdins5P. Here, by demonstrating AtMTM phospholipid 3′-phosphatase activity, its substrate specificity, and its increased expression under water withdrawal (microarray data and Fig. 3B–E), it is conceivable that production of endogenous Ptdins5P was elevated in OX-MTM and in dehydrated wild type cells.
Collectively, the biochemical and the microarray data reported in the paper are consistent with AtMTM1 and ATX1 participating in a partially overlapping drought-response pathway and that overproduction of AtMTM1 negatively affects ATX1 activity, most likely through the lipid. Another intriguing relationship between myotubularins and Trithorax proteins relevant is the ability of a conserved domain (SID) found in all myotubularins to specifically bind the SET-domain (Set interacting domain).40 The biological significance of this interaction is debated, as crystallographic studies have shown that the SID peptide is buried inside the tertiary structure of the protein suggesting that SID is involved in stabilizing myotubularin structure rather than in binding with other proteins.33 However, mutations in SID aborting its binding with SET resulted in abnormal growth and leukemia.40,41 Furthermore, the inactive myotubularin, Sbf1, was isolated as a component of the Trx complex, physically and functionally linked with the trithorax protein in Drosophila.42 An important implication of these results is that myotubularins, active or inactive, may have nuclear localization and could participate in chromatin remodeling.43 A variety of modifying enzyme activities, including lipases, lipid kinases and lipid phosphatases have been found at the nuclear membrane and the nuclear matrix.44,45 It remains to be shown whether an AtMTM might be present also in the nucleus.
Materials and Methods
Plant material: Stressing plants by water withdrawal.
Arabidopsis Col wild type, atx1 mutant, and OX-AtMTM seeds were grown in soil at 22°C under a cycle of 14 hr light/10 hr darkness. At day 24, the samples for drought treatment were deprived of irrigation for three days, followed by three additional days during which the plants, with the soil, were taken out of the pot for the day but placed back into the pots during the night with constant monitoring of the water loss. Tissues for microarray analyses were collected when final relative water concentration (RWC) was 65%. RWC was measured according to the formula:
RWC = (FW - DW)/(SW - DW) × 100%
as follows: detached leaves were weighted as fresh weight (FW) followed by submergence in water overnight in 4°C to measure saturated weight (SW). The samples were dried at 65°C for 24 h to measure the dry weight (DW). Non-stressed samples for wild type, atx1 and OX-AtMTM were grown under the same conditions but with regular irrigation.
Recombinant AtMTM protein and generation of stably overexpressing transgenic lines.
Mining the public databases with the sequence for the human MTMR2 as a probe, we identified a cDNA for At3g10550 (RAFL15-11-P18), which was obtained from RIKEN (Japan). In this clone, there was a stop codon at position 1,580 bp producing a truncated protein. However, a partial cDNA clone overlapping this area in the correct frame (NCBI acc. #BX839151 EST) was obtained from INRA (France). After a series of cloning steps, we generated a clone containing the entire AtMTM1 sequence, as well as various deletion forms. The DNA sequence was verified by sequencing. The protein containing the catalytic PTP and the adjacent RIC and SID domains was cloned in the pGEX4T-1 vector (Amersham), expressed as a GST-fusion in E. coli BL21 cells, and used to assay the enzyme activity.
For generating stably AtMTM-overexpressing lines, the full-length sequence was amplified by PCR specific primers containing the attB1 and attB2 sequences, and recombined by the attB × attP (BP) reaction into pDONR221 (Invitrogen). pDONR221/AtMTM1 was recombined into vectors by the LR reaction.29 The binary vector pAlligator2-AtMTM construct containing the coding sequence of the full length AtMTM gene and driven by the 35S promoter was used for transformation in Arabidopsis.30 The binary vector contains an N-terminal HA-fusion tag and a selectable marker (GFP driven by a seed coats-specific promoter) facilitating isolation of transformed seeds.
Phosphatase activity.
Phosphoinositide 3-phosphatase assays were performed by the malachite green assay.28,31 Recombinantly expressed and affinity-column purified GST-tagged proteins were reacted with the substrates (2.5 µmoles per experimental point) in 50 µl assay buffer (50 mM Tris/HCl, pH 8.0, 100 mM KCl and 2 mM DTT). Mono- and Di-C8 phosphoinositides (Echelon Biosciences Inc.,) were used as substrates and PTEN (phosphatase and tensin) lipid phosphatase (Echelon Biosciences Inc., E-3000) was used as a positive control. Following incubation at 37°C for 30 min, the reactions were quenched by the addition of 20 µl of 0.1 M N-ethylmaleimide and spun at 18,000 g for 10 min. Twenty-five microliters of the supernatant was added to 100 µl of malachite green reagent and vortexed. Samples were allowed to sit for 40 min for color development before measuring absorbance at 620 nm. Inorganic phosphate release was measured by a standard curve of KH2PO4 in distilled water.
RNA sample and microarray preparation.
In two independent experiments, RNAs were isolated from control watered, and from drought-stressed plants grown under the same conditions. Total RNA was extracted from frozen plants using TRIzol reagent following the manufacturer's instructions (Invitrogen) and further purified using Qiagen RNeasy column (Qiagen). Total RNA was used to synthesize cDNA using Affymetrix One-Cycle cDNA Synthesis Kit according to the manufacturer's instructions (Affymetrix). All sample preparations followed prescribed protocols (Affymentrix GeneChip Transcript levels Analysis Technical manual). Hybridization was done on an Affymetrix Arabidopsis Genome ATH1 Array, stained with streptavidin-phycoerythrin conjugate on an Affymentrix Fluidics Station 450, followed by scanning with the GeneChip Scanner 3000 7G (Affymetrix). Affymetrix GeneChip Operating Software (GCOS) was used for washing, scanning, and basic data analysis.
Microarray data analyses.
Affymetrix gene chips carrying 22,500 probe sets (∼24,000 Arabidopsis genes) were used for whole-genome transcript profile analysis of atx1 and OX-AtMTM lines, as well as wild type Col plants under non-stress (irrigated) and upon water withdrawal (stress) conditions. Patterns reflect whole-plant gene transcript levels profiles at this developmental stage. Experiments were performed in separate duplicate hybridizations with two independently isolated RNA samples. Signal variations between two replicate samples for each gene is reflected by the correlation coefficients.
A data-mining tool developed by Lu et al.32 was used to identify genes significantly expressed in mutant and wild type samples. Data analysis by GCOS is performed in two steps: single array analysis and comparison analysis. First, data were processed for background correction, value extraction, data summarization and normalization. The One-Step Tukey's biweight method is used to estimate the variance among different probe pairs within a probe set and to produce a single transcript levels index that represented transcript abundance. The One-Sided Wilcoxon signed-rank test is used to generate the Detection p-values (P, p < 0.04) or (A, p > 0.06). Array normalizations were preformed by the scaling approach, which adjusts the average intensity of every array to a common value in order to make the arrays comparable. Hybridization data in replicate experiments showed correlation coefficients ranging from 0.86 to 0.97. The cutoff for the log2 signal ratio is 1.5 corresponding to ∼three-fold change in signal intensity levels between the experimental and control data sets.
Cluster (Overlap) analyses are performed in all possible configurations to identify common genes regulated by the tested factors. Results are presented as Venn diagrams, illustrating the overlaps, and in tables, identifying individual shared genes.
Significance analysis for the overrepresentation of membrane and wall-associated genes was done by two tools as to get two different opinions: via AmiGO and GOEAST. The least significant p value of both tools was taken as a very conservative indication of the significance. p values for the membrane and wall associated genes shared between atx1/wt-stressed and between atx1/MYO-OX/wt stressed genomes was p ≤ 0.000006 and p ≤ 0.005, respectively.
qRT-PCR analysis.
RNA for the qRT-PCR (quantitative realtime reverse transcription-polymerase chain reaction) was isolated with TRIZOL (Invitrogen) and purified with RNasy plant mini kit (Qiagen, #74903). For the first strand cDNA synthesis, 8 µg total RNA was treated with DNase I, extracted with phenol and chloroform, precipitated with ethanol, followed by addition of oligo (dT) and superscript III reverse transcriptase (Invitrogen). Real-time PCR analysis was performed using the iCycleriQ realtime PCR instrument (Bio-Rad) and iQ SYBR Green Supermix (BioRad). The relative accumulation of transcripts from specific genes was quantitated using 2−ΔΔCt calculation, where ΔΔCt is the difference in the threshold cycles of the test and housekeeping gene ubiquitin. ΔΔCt is the threshold cycle of the target gene subtracted from the threshold cycle of the housekeeping gene. The mean threshold cycle values for the genes of interest were calculated from three experiments. Used primers:
Ubiquitin: F-(5′-AGG ATG GCA GAA CTC TTG CT-3′), R-(5′-TCC CAG TCA ACG TCT TAA CG-3′); At3g02480, F-(5′-TGC AAC AGA CTG GAC AAC AA-3′), R-(5′-TCC AAG GTG CTT GCT TAG TG-3′); At3g17520, F-(5′-AGA TAA GGC GTC GCA GAG TT-3′), R-(5′-ATC GTC GGT CAA GCT CTC TT-3′), At1g13260, F-(5′-ATA CCG AAA CAT CAC GCA GA-3′), R-(5′-AAC GGA ACC TCC ACA CTT TC-3′), At1g21270, F-(5′-TTC TCG GTT ATC ATG CTT GG-3′), R-(5′-CTC GCT GTA TCA ACA TGC CT-3′), At1G29430, F-(5′-ACG GCT GAT AAG ATC CGT TT-3′), R-(5′-GAA TGG CAA TGT GAT TGG TC-3′), At1g75250, F-(5′-GAA GTG AAG CGC CAC TAT GA-3′), R-(5′-GTA ATT GGG CAA AGG GAC AC-3′), At5g44020, F-(5′-AGG TGG TTC CAC AGG AAT GT-3′), R-(5′-GCA TGT CTT CTT CTC GCA AC-3′), At1g29660, F-(5′-GCA GAC AGT ACA CTC CCG AA-3′), R-(5′-TCC AAT TCC TAC CAA TGC AA-3′), At2g10940, F-(5′-CAG TCC CTA AGC TAC CCG TT-3′), R-(5′-GAG GTA TGG GTA GCC CTG AA-3′), At2g21220, F-(5′-CAA GAG CTT GCT TCA ACA GG-3′), R-(5′-TGG ATG TTA AAG AGC GGA AA-3′), ACTIN7, F-(5′-CTA CGA GGG GTA TGC TCT TCC TCA T-3′), R-(5′-CTG AAG AAC TGC TCT TGG CTG TCT C-3′), ATX1, F-(5′-CCC AAT TGC TAT TCT CGA GTC ATC A-3′), R-(5′-TTT GCA TGT TGT TCT TCA GCT TCT G-3′), AtMTM, F-(5′-CCC AAG GAG CTC TCT GGA GAA TAA C-3′), R-(5′-CTT CCG ACA TGA GCA CAT CCT ACT T-3′).
Supplementary Material
Acknowledgements
This work was partially supported by NSF-MCB0749504 award to Z.A. and NSF-EPS0701892 award to M.F. and Z.A.
Footnotes
Previously published online: www.landesbioscience.com/journals/psb/article/10103
References
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:1–4. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Grimaud C, Negre N, Cavalli G. From genetics to epigenetics: the tale of Polycomb group and trithorax group genes. Chromosome Res. 2006;14:363–375. doi: 10.1007/s10577-006-1069-y. [DOI] [PubMed] [Google Scholar]
- 3.Kiefer JC. Epigenetics in development. Dev Dyn. 2007;236:1144–1156. doi: 10.1002/dvdy.21094. [DOI] [PubMed] [Google Scholar]
- 4.Bird A. Perceptions of epigenetics. Nature. 2007;447:396–398. doi: 10.1038/nature05913. [DOI] [PubMed] [Google Scholar]
- 5.Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- 6.Pien S, Grossniklaus U. Polycomb group and trithorax group proteins in Arabidopsis. Biochim Biophys Acta. 2007;1769:375–382. doi: 10.1016/j.bbaexp.2007.01.010. [DOI] [PubMed] [Google Scholar]
- 7.Álvarez-Venegas R, Al-Abdallat A, Guo M, Alfano JP, Avramova Z. Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics. 2007;3:106–113. doi: 10.4161/epi.2.2.4404. [DOI] [PubMed] [Google Scholar]
- 8.Van den Burg HA, Takken FLW. Does chromatin remodeling mark systemic acquired resistance? Trends Plant Sci. 2009;14(286) doi: 10.1016/j.tplants.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Álvarez-Venegas R, Sadder M, Hlavacka A, Baluška F, Xia Y, Lu G, et al. The Arabidopsis Homolog of Trithorax, ATX1, Binds Phosphoinositide 5-Phosphate and the Two Regulate a Common Set of Target Genes. Proc Natl Acad Sci. 2006;103:6049–6054. doi: 10.1073/pnas.0600944103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim JM, To TK, Ishida J, Morosawa T, Kawashima M, Matsui A, et al. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Phys. 2008;49:1580–1588. doi: 10.1093/pcp/pcn133. [DOI] [PubMed] [Google Scholar]
- 11.Bergman A, Siegal ML. Evolutionary capacitance as a general feature of complex gene networks. Nature. 2003;424:549–552. doi: 10.1038/nature01765. [DOI] [PubMed] [Google Scholar]
- 12.Lee I, Lehner B, Crombie C, Wong W, Fraser AG, Marcotte EM. A single gene network accurately predicts phenotypic effects of gene perturbation in Caenorhabditis elegans. Nat Genet. 2008;40:181–188. doi: 10.1038/ng.2007.70. [DOI] [PubMed] [Google Scholar]
- 13.Lehner B, Crombie C, Tischler J, Fortunato A, Fraser AG. Systematic mapping of genetic interactions in Caenorhabditis elegans identifies common modifiers of diverse signaling pathways. Nat Genet. 2006;38:896–903. doi: 10.1038/ng1844. [DOI] [PubMed] [Google Scholar]
- 14.Álvarez-Venegas R, Avramova Z. Methylation Patterns of Histone H3 Lys4, Lys 9 and Lys 27 in transcriptionally active and inactive Arabidopsis genes and in atx1 mutants. Nucl Acids Res. 2005;33:5199–5207. doi: 10.1093/nar/gki830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Saleh A, Alvarez-Venegas R, Yilmaz M, Le O, Hou G, Sadder M, et al. The Highly Similar Arabidopsis Homologs of Trithorax ATX1 and ATX2 Encode Proteins with Divergent Biochemical Functions. Plant Cell. 2008;20:568–579. doi: 10.1105/tpc.107.056614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Álvarez-Venegas R, Xia Y, Lu G, Avramova Z. Phosphoinositide 5-phosphate and phosphoinositide 4-phosphate trigger distinct specific responses of Arabidopsis genes; genome-wide transcript levels analyses. Plant Signal Behavior. 2006;1:140–151. doi: 10.4161/psb.1.3.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dove SK, Cooke FT, Douglas MR, Sayers LG, Parker PJ, Michell RH. Osmotic stress activates phosphatidyl-D-inositol 3,5-bisphosphate synthesis in yeast. Nature. 1997;190:187–192. doi: 10.1038/36613. [DOI] [PubMed] [Google Scholar]
- 18.Blondeau F, Laporte J, Bodin S, Superti-Furga G, Payrastre B, Mandel JL. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum Mol Genet. 2000;9:2223–2229. doi: 10.1093/oxfordjournals.hmg.a018913. [DOI] [PubMed] [Google Scholar]
- 19.Sbrissa D, Ikonomov O, Deeb R, Shisheva A. Phosphatidylinositol 5-phosphate biosynthesis is linked to PIKfyve and is involved in osmotic response pathway in mammalian cells. J Biol Chem. 2002;277:47276–47284. doi: 10.1074/jbc.M207576200. [DOI] [PubMed] [Google Scholar]
- 20.Meijer HJG, Berrier CP, Iurisci C, Divecha N, Musgrave A, Munnik T. Hyperosmotic stress induces rapid synthesis of phosphatidyl-D-inositol 3,5-bisphosphate in plant cells. Biochem J. 2001;360:491–498. doi: 10.1042/0264-6021:3600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pical C, Westergren T, Dove SK, Larsson, Sommarin M. Salinity and hyperosmotic stress induce rapid increases in phosphatidylinositol 4,5-bisphosphate, diacylglycerol phosphate and phosphatydylcholine in Arabidopsis thaliana cells. J Biol Chem. 1999;274:38232–38240. doi: 10.1074/jbc.274.53.38232. [DOI] [PubMed] [Google Scholar]
- 22.Berger P, Bonneick S, Willi S, Wymann M, Suter U. Loss of phosphatase activity in myotubularin-related protein 2 is associated with Charcot-Marie-Tooth disease type 4B1. Hum Mol Genet. 2002;15:1569–1579. doi: 10.1093/hmg/11.13.1569. [DOI] [PubMed] [Google Scholar]
- 23.Lorenzo O, Urbé S, Clague MJ. Analysis of phosphoinositide binding domain properties within the myotubularin-related protein MTMR3. J Cell Sci. 2005;118:2005–2012. doi: 10.1242/jcs.02325. [DOI] [PubMed] [Google Scholar]
- 24.Kim SA, Taylor GS, Torgersen KM, Dixon JE. Myotubularin and MTMR2, phosphatidylinositol 3-phosphatases mutated in myotubular myopathy and type 4B Charcot-Marie-Tooth disease. J Biol Chem. 2002;277:4526–4531. doi: 10.1074/jbc.M111087200. [DOI] [PubMed] [Google Scholar]
- 25.Clague MJ, Lorenzo O. The myotubularin family of lipid phosphatases. Traffic. 2005;6:1–7. doi: 10.1111/j.1600-0854.2005.00338.x. [DOI] [PubMed] [Google Scholar]
- 26.Pendaries C, Tronchere H, Racaud-Sultan C, Gaits-Iacovoni F, Coronas S, Manenti S, et al. Emerging roles of phosphatidyl inositol monophosphates in cellular signaling and trafficking. Adv Enzyme Reg. 2005;45:201–204. doi: 10.1016/j.advenzreg.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 27.Robinson FL, Dixon J. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 28.Schaletzky J, Dove SK, Short B, Lorenzo O, Clague MJ, Barr FA. Phosphatidylinositol-5-phosphate activation and conserved substrate specificity of the myotubularin phosphatidylinositol 3-phosphatases. Curr Biol. 2003;13:504–509. doi: 10.1016/s0960-9822(03)00132-5. [DOI] [PubMed] [Google Scholar]
- 29.Karimi M, Inze D, Depicker A. Gateway vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002;7:193–195. doi: 10.1016/s1360-1385(02)02251-3. [DOI] [PubMed] [Google Scholar]
- 30.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 31.Martin B, Pallen CJ, Wang JH, Graves DJ. Use of fluorinated tyrosine phosphates to probe the substrate specificity of the low molecular weight phosphatase activity of calcineurin. J Biol Chem. 1985;260:14932–14937. [PubMed] [Google Scholar]
- 32.Lu G, Nguyen TV, Xiao Y, Fromm M. AffyMiner: mining differentially expressed genes and biological knowledge in GeneChip microarray data. BMC Bioinformatics. 2006;7:26. doi: 10.1186/1471-2105-7-S4-S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Begley MI, Dixon JE. The structure and regulation of myotubularin phosphatases. Curr Opin Struct Biol. 2005;15:614–620. doi: 10.1016/j.sbi.2005.10.016. [DOI] [PubMed] [Google Scholar]
- 34.Begley MJ, Taylor GS, Brock MA, Ghosh P, Woods VL, Dixon JE. Molecular basis for substrate recognition by MTMR2, a myotubularin family phosphoinositide phosphatase. Proc Natl Acad Sci. 2006;103:927–932. doi: 10.1073/pnas.0510006103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Álvarez-Venegas R, Pien S, Sadder M, Witmer X, Grossniklaus U, Avramova Z. ATX-1, an Arabidopsis homolog of trithorax, activates flower homeotic genes. Curr Biol. 2003;13:627–637. doi: 10.1016/s0960-9822(03)00243-4. [DOI] [PubMed] [Google Scholar]
- 36.Qin F, Sakuma Y, Tran LS, Maruyama K, Kidokoro S, Fujita Y, et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene transcript levels. Plant Cell. 2008;20:1693–1707. doi: 10.1105/tpc.107.057380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yamaguchi-Shinozaki K, Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stress. Annu Rev Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 38.Ndamukong I, Chetram A, Saleh A, Avramova Z. Wall-modifying genes regulated by the Arabidopsis homolog of trithorax, ATX1; repression of the XTH33 gene as a test case. Plant J. 2009;58:541–553. doi: 10.1111/j.1365-313X.2009.03798.x. [DOI] [PubMed] [Google Scholar]
- 39.Tronchere H, Laporte J, Pendaries C, Chaussade C, Liaubet L, Pirola L, et al. Production of phosphatidylinositol 5-phosphate by the phosphoinositide 3-phosphatase myotubularin in mammalian cells. J Biol Chem. 2004;279:7304–7312. doi: 10.1074/jbc.M311071200. [DOI] [PubMed] [Google Scholar]
- 40.Cui X, De Vivo I, Slany R, Miyamoto A, Firestein R, Claery ML. Association of SET domain and myotubularin-related proteins modulates growth control. Nat Genet. 1998;18:303–305. doi: 10.1038/ng0498-331. [DOI] [PubMed] [Google Scholar]
- 41.Firestein R, Cui X, Huie P, Claery ML. Set domain dependent regulation of transcriptional silencing and growth control by SUV39H1, a mammalian ortholog of Drosophila Su(var)3–9. Mol Cell Biol. 2000;20:4900–4909. doi: 10.1128/mcb.20.13.4900-4909.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Petruk S, Sedkov Y, Smith S, Tillib S, Kraevski V, Nakamura T, et al. Trithorax and dCBP acting in a complex to maintain transcript levels of a homeotic gene. Science. 2001;294:1331–1334. doi: 10.1126/science.1065683. [DOI] [PubMed] [Google Scholar]
- 43.Firestein R, Cleary ML. Pseudo phosphatase Sbf1 contains an N-terminal GEF homology domain that modulates its growth regulatory properties. J Cell Sci. 2001;114:2921–2927. doi: 10.1242/jcs.114.16.2921. [DOI] [PubMed] [Google Scholar]
- 44.Irvine RF. Nuclear lipid signaling. Nature Rev. 2003;4:1–12. doi: 10.1038/nrm1100. [DOI] [PubMed] [Google Scholar]
- 45.Jones DR, Divecha N. Linking lipids to chromatin. Curr Opin Genet Develop. 2004;14:196–202. doi: 10.1016/j.gde.2004.02.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.