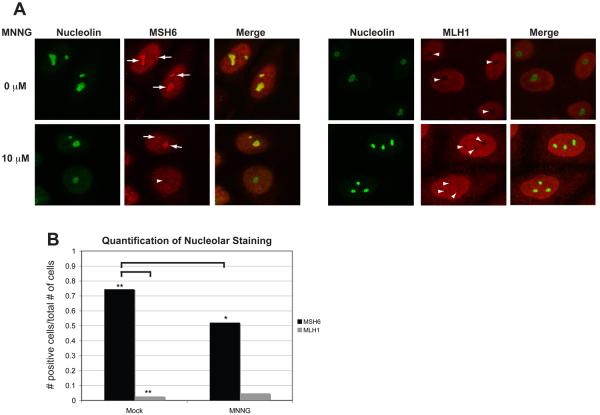
Fig. 8.
A population of hMSH6, but not hMLH1, localizes to the nucleolus in the absence of DNA damage. (A) HeLa cells were plated onto glass coverslips, synchronized by a DTB and treated with 10 μM MNNG and O6-BG or DMSO. Coverslips were removed at 4 hours post-release and immediately processed for confocal immunofluorescence with antibodies against hMSH6, hMLH1, and Nucleolin. A population of hMSH6 can be detected within nucleoli as marked by positive nucleolin staining in mock-treated cells. The nucleolar localization is dramatically reduced in MNNG-treated cells. hMLH1 rarely appears to localize within the nucleoli. (B) Quantification of nucleolar staining. >150 cells for each condition were counted. p<0.0001 when comparing hMSH6 mock versus hMSH6 MNNG by a paired T-test. p< 0.0001 when comparing hMSH6 mock versus hMLH1 mock by a paired T-test.