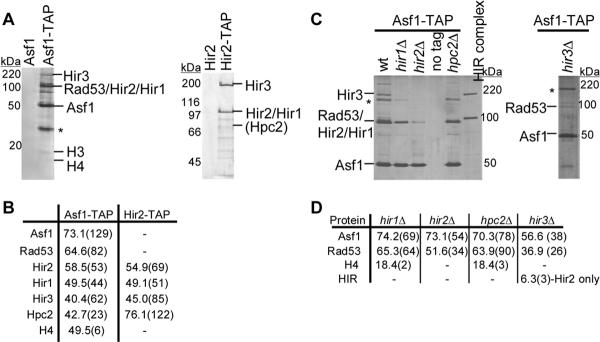
Figure 1.
The four Hir proteins exist as a complex in vivo and co-purify with Asf1. (A) TAP purifications were performed using wild-type (PKY028) and ASF1-STAP (PKY3121) cells (left panel) and from wildtype (PKY028) and HIR2-STAP cells (PKY3062) (right panel). The Asf1-STAP purification was performed at 100 mM KCl and the Hir2-STAP purification was performed at 300 mM KCl. Proteins eluted from the second TAP affinity resin were analyzed on a SDS-PAGE gel and detected by silver staining. Polypeptides identified by mass spectrometry, immunoblotting or molecular weight (histone H3) are indicated. *indicates an unidentified polypeptide. (B) Summary of Asf1-STAP and Hir2-STAP mass spectrometry data. The percent sequence coverage is indicated in the table, with the number of unique peptides shown in parentheses. Although histone H3 peptides were not detected in all Asf1-STAP preparations by mass spectrometry, H3 was always apparent on the silver-stained protein gels. (C) Efficient association between Asf1 and the HIR complex is dependent upon all four Hir proteins. Asf1-TAP complexes were purified from wildtype (PKY3121), hir1Δ (PKY3906), hir2Δ (PKY3908), hpc2Δ (PKY3912) and hir3Δ (PKY3928) cells, analyzed on SDS-PAGE gels and detected by silver staining. Purified Hir2-STAP complex is shown for comparison. The Hir2-STAP subunit migrates more slowly than Rad53, which is still present in the complexes from hir mutant cells. *indicates an unidentified polypeptide. (D) A summary of Asf1-TAP mass spectrometry data. The percent sequence coverage of each protein is shown, with the number of unique peptides in parentheses.