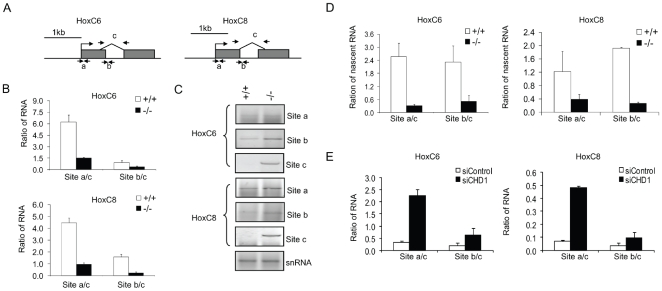
Figure 5. Enhanced transcriptional elongation and generation of spliced mature transcripts after Lsh depletion.
(A) Schematic representation of the probe and primer pairs used to detect 5′ primary transcripts, unspliced transcripts and spliced mRNA. (B) Real time RT-PCR analysis detecting the ratio of unspliced to spliced mRNA of the HoxC6 and HoxC8 gene from Lsh+/+ and Lsh−/− MEFs. Results represent the mean and standard deviations of three independent experiments. (C) RT-PCR analysis for detection of HoxC6 and HoxC8 transcripts derived from nuclei of Lsh+/+ and Lsh−/− MEFs. U2 small nuclear RNA (snRNA) was unaffected by Lsh and served as a control. (D) Real-time RT-PCR analysis from four independent experiments to detect the ratio of unspliced to spliced nascent transcripts of HoxC6 and HoxC8 genes by nuclear run on assay. (E) Real time RT-PCR analyses detecting the ratio of unspliced to spliced mRNA of the HoxC6 and HoxC8 genes in Lsh−/− MEFs after Chd1 siRNA treatment. Error bars represent the standard deviations for the mean of three independent experiments.