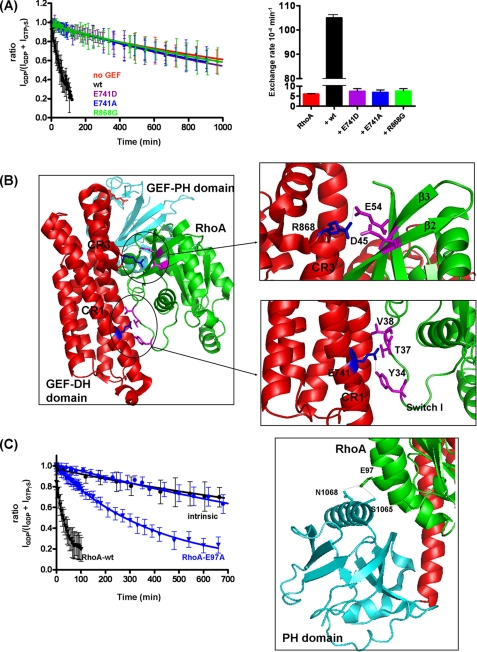
FIGURE 4.
Mutations in conserved regions of DH-PHPRG severely decrease GEF activity, and E97A mutation decreases the activation of RhoA. A, RhoA nucleotide exchange (GDP to GTPγS) stimulated by wild-type (wt) DH-PHPRG (1.8 nm) versus mutants E741D, E741A, and R868G. Rates derived from these curves are displayed in a histogram in the right panel. The experiments were also performed with 4 nm DH-PHPRG, in which wild-type, E741D, E741A, R868G, and intrinsic nucleotide exchange rates are 207 × 10−4 ± 3.3 × 10−5 min−1, 6.4 × 10−4 ± 1 × 10−4 min−1, 14.4 × 10−4 ± 1 × 10−4 min−1, 11.1 × 10−4 ± 1 × 10−4 min−1 and 4.4 × 10−4 ± 2 × 10−4 min−1, respectively. B, interface of PDZ-RhoGEF DH (red) and PH domains (cyan) with RhoA (green). DH-PHPRG Glu-741 (in CR1), Arg-868 (near CR3), and RhoA switch I residues Tyr-34, Thr-37, and Val-38 are highlighted (PDB code: 1XCG). PyMOL software was used to create the schematic representations. C, mutation of Glu-97 reduces the ability of DH-PHPRG to activate RhoA. RhoA nucleotide exchange to GTPγS in the absence (circles) or presence (triangles) of DH-PHPRG (4 nm). Black and blue curves correspond to wild-type RhoA and RhoA-E97A, respectively. Right panel, RhoA Glu-97 forms hydrogen bonds with Ser-1065 and Asn-1068 of the PH domain. In A and C, error bars indicate S.D. for values reported by multiple peaks.