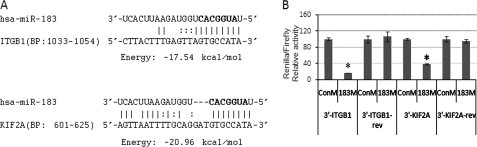
FIGURE 1.
Targeting of the 3′-UTRs of ITGB1 and KIF2A by miR-183. A, potential binding sites for miR-183 in the 3′-UTRs of ITGB1 and KIF2A predicted by computational analysis. BP indicates base pairs of the 3′-UTR of each gene where the sequences are located. The seed regions are highlighted in bold. B, to test for potential interactions with miR-183, the complete 3′-UTRs of ITGB1 (3′-ITGB1) and KIF2A (3′-KIF2A) were cloned into the psiChECK2 dual-luciferase reporter vector and co-transfected with either miR-183 mimic (183M) or scramble control mimic (ConM) into human embryonic kidney 293 cells. Negative controls were generated by cloning the same 3′-UTRs in reverse orientation (3′-ITGB1-rev and 3′-KIF2A-rev). The luciferase activities of Renilla (with inset) and firefly (endogenous) were analyzed 24 h post-transfection. The data represent the percentage of changes in the ratio of Renilla/firefly activities compared with the controls ± S.D. (n = 3; *, p < 0.05 by Mann-Whitney U test). Error bars are S.D.