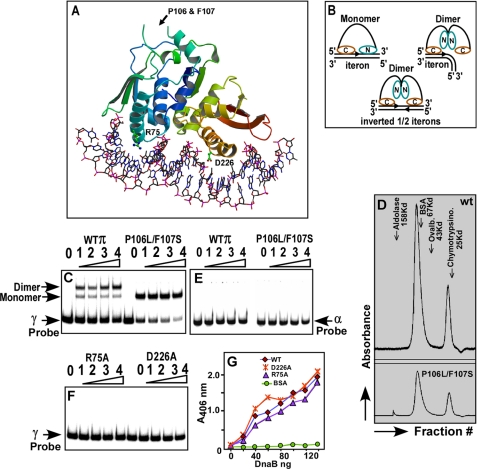
FIGURE 2.
Structure of the monomeric π iteron complex and the postulated configuration of the dimeric and monomeric π protein bound to iteron DNA. A, crystal structure of monomeric π-iteron complex showing the two DNA recognition α helices; the N terminus is blue-green and the C terminus is red-brown. The locations of the various mutations used in this work are shown, and the structure is a winged helix. B, models based on footprinting data and crystal structure showing the postulated binding of monomeric and dimeric π to the iterons and the binding of dimeric π to the inverted half-iterons present at the π operator sequence shown in Fig. 1A; note that the dimer is postulated to undergo a structural alteration that prevents the N-terminal recognition helix from binding to the 3′-half of a full iteron sequence. C, gel mobility shift experiments showing that the dimer binds to a labeled γ iteron, giving a major shift corresponding to the dimer and a minor one corresponding to the monomer. The monomeric πP106L F107S (called π cop) binds to the γ iteron probe and exclusively generates a band corresponding to a monomeric π iteron complex; note that π cop binds with higher affinity to the DNA than the wt π as revealed by the intensity of the unbound probe. D, gel filtration profiles of the dimeric wt π (upper panel) and π cop (lower panel)) with marker proteins, showing a majority of dimeric protein (with a lesser peak corresponding to the monomer). E, neither the wt π nor the π cop binds to the α iteron probe under conditions identical to that shown in C. F, gel mobility shift experiments showing that the R75A and D226A mutants fail to bind to DNA. In all gel mobility shift experiments, 20 fmol of labeled DNA were incubated with 0, 8, 16, 32, and 64 pmol of π (wt, cop, R75A, and D226A forms). G, ELISA showing the binding of DnaB to various forms of purified π; note that although the R75A and D226A mutant forms are completely defective in iteron binding, these are almost as effective as wt π in binding to DnaB.