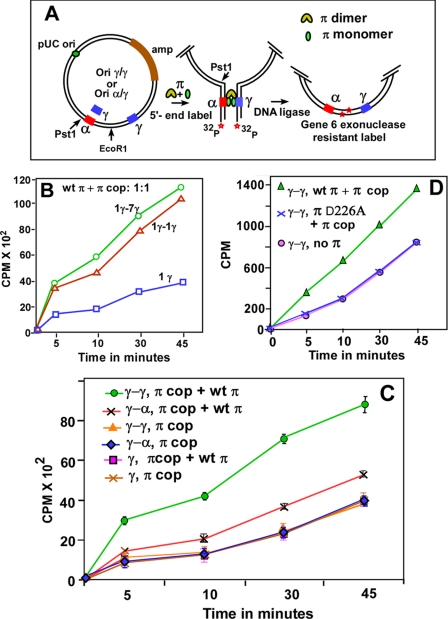
FIGURE 3.
Determination of the frequency of DNA looping by enhancement of ligase-catalyzed DNA circularization. A, schematic depiction of the circularization assay showing the pUC19-based substrate that contained γ-γ or α-γ iterons cloned at the indicated locations on either side of a unique EcoR1 site. A similar plasmid having a single γ iteron on one side and the 7 γ iterons on the other was also constructed. The assay steps including linearization by EcoR1, 5′-end labeling, incubation with either π cop, or with a 1:1 mixture of wt π and π cop, ligation with T4 DNA ligase at 16 °C for various periods of time, and digestion with T7 gene 6 exonuclease are shown; 20 fmol of DNA were incubated with a total of 128 pmol of π. B, DNA circularization kinetics showing that there was a small difference between γ-γ and γ-7 γ iteron interactions; the control experiment with a solo γ iteron is also shown. C, DNA circularization kinetics with various combinations of DNA substrates and π proteins as indicated. The α-γ plasmid incubated with a 1:1 mixture of wt π and π cop had a small but distinct and reproducible enhancement in ligation kinetics over the same incubated with π cop only. The γ-γ plasmid incubated with π monomer-dimer mixture had a much higher rate of DNA circularization over that when only π cop was used. Data were collected from six separate sets of experiments and are presented with the S.D. as error bars. D, ligation experiments showing that π D226A (non-DNA binding mutant dimers) did not complement π cop to loop two γ-γ iterons. Controls show that under identical conditions, wt π complemented π cop in promoting γ-γ interaction, and there was no interaction as expected when π was withheld from the reaction.