Abstract
In Escherichia coli, expression of the RelE and HipA toxins in the absence of their cognate antitoxins has been associated with generating multidrug-tolerant “persisters.” Here we show that unlike persisters of E. coli, persisters of Mycobacterium tuberculosis selected with one drug do not acquire cross-resistance to other classes of drugs. M. tuberculosis has three homologs of RelE arranged in operons with their apparent antitoxins. Each toxin individually arrests growth of both M. tuberculosis and E. coli, an effect that is neutralized by coexpression of the cognate antitoxin. Overexpression or deletion of each of the RelE toxins had a toxin- and drug-specific effect on the proportion of bacilli surviving antibiotic killing. All three toxins were upregulated in vivo, but none of the deletions affected survival during murine infection. RelE2 overexpression increased bacterial survival rates in the presence of rifampin in vitro, while deletion significantly decreased survival rates. Strikingly, deletion of this toxin had no discernible effect on the level of persisters seen in rifampin-treated mice. Our results suggest that, in vivo, RelE-generated persisters are unlikely to play a significant role in the generation of bacilli that survive in the face of multidrug therapy or in the generation of multidrug-resistant M. tuberculosis.
Chemotherapy of tuberculosis requires 2 months of treatment with a combination of four antituberculosis drugs, isoniazid (INH), rifampin (Rif), pyrazinamide (PZA), and ethambutol, followed by at least 4 months of continuing therapy with INH and Rif (48). Although sterilization of the sputum of most patients can be achieved in 2 months, an additional 4 to 7 months is necessary to reduce relapse to acceptable levels (43). The extended duration of chemotherapy required to combat relapse following chemotherapy seen with pulmonary tuberculosis has long been attributed to persistent bacilli (17, 39, 40). One hypothesis is that subpopulations of bacilli reside in distinct physical niches in which only certain drugs are active because of adaptive alterations in the metabolism of the pathogen (4). It has been hypothesized that these bacilli are metabolically inactive due to orchestrated alterations in microbial metabolism in response to hypoxia, nitrosative stress, and/or nutrient deprivation (10, 51, 56) and that the bacilli are drug tolerant by virtue of their lowered or altered metabolism (19, 40). Another hypothesis is that subpopulations of bacilli in identical physical environments are in stochastically generated heterogeneous metabolic states, some of which are antibiotic tolerant (57).
Both hypotheses predict the existence of drug-tolerant “persister” bacilli whose sterilization rate is associated with the length of time required to achieve a durable cure of tuberculosis. Persisters, in the sense most relevant to shortening chemotherapy, are bacilli that remain after months of chemotherapy. If chemotherapy is truncated, these remaining bacilli remain capable of reactivating metabolism to induce relapse with active disease, a phenomenon that can be modeled somewhat in mouse models (21, 36-38, 50). Reproducibility, the extensive duration of the experiments, the relevance of the murine model to human disease, and the lack of observable bacilli have severely limited the utility of such models (51, 56).
Bacterial drug tolerance is reported to result from lower metabolic requirements for processes that characterize actively growing cells, such as transcription, translation, replication, and cell wall synthesis (26, 27, 53, 54). Such processes are typical targets for antibiotics, and cells that are not actively replicating are therefore thought to be broadly tolerant to multiple drugs and antibiotics. This correlation of metabolic activity with sensitivity to drugs has been suggested by a variety of experimental findings, including (i) the growth rate dependence of killing of cells by several antibiotics (15), (ii) the increased number of persisters associated with ectopic expression of proteins that directly inhibit processes in macromolecular synthesis (27), and (iii) the reduction in the number of persisters in cells that are defective in regulating nutrient transport, energy metabolism, and other aspects of active metabolism (35). In fact, isolation of cells with low metabolic rates revealed a transcriptionally discrete subpopulation highly enriched with drug-tolerant bacteria (52).
An attractive hypothesis for the origin of these persisters is that they arise from the stochastic overexpression of endogenous regulators of macromolecular synthesis in a subset of cells (27, 52, 53). The best studied of these systems are the “toxin-antitoxin” (TA) modules found within many prokaryotic genomes (44). These modules are generally expressed from a bicistronic operon wherein the upstream gene encodes an unstable antitoxin and the downstream gene encodes a stable toxin (18, 22). The antitoxins neutralize their cognate toxins by forming tight protein-protein complexes that abrogate toxicity if both modules are present in equal concentrations (25). The two proteins have differing stabilities, however, so that in the absence of continued expression, the unstable antitoxin is eventually degraded, leading to growth arrest by the toxin (8).
Many distinct TA families that impinge on persister formation have been described, including HipBA, MazEF, RelBE, CcdAB, ParDE, HigBA, Phd/doc, and VapBC (22, 44). This effect was first described for mutations in hipA, which increase the proportion of persisters, with up to 10% of bacilli surviving drug treatment (30, 31, 41). Likewise, RelE or MazF overexpression increases the number of persisters following antibiotic treatment (27, 33). Persisters that survive drug treatment exhibit distinct transcriptional signatures. In one study of Escherichia coli, it was observed that at least 300 genes were differentially expressed in a persister population, 2% of which comprised TA modules, including dinJ yafQ, yefM, relBE, and mazEF (27).
RelE leads to bacteriostasis by cleaving of the coding region of mRNAs at the ribosomal A site, resulting in ribosomal stalling on the truncated message (20, 22, 46). Overexpression of transfer-messenger RNA (tmRNA) releases stalled ribosomes from the transcript and tags the truncated peptides for proteolytic degradation (9, 24). The expression of the relBE operon is minimal under normal logarithmic growth conditions, but stresses, such as amino acid starvation, lead to rapid depletion of RelB and growth arrest by RelE (8, 34). We have previously demonstrated that treatment of Mycobacterium tuberculosis with a variety of drugs resulted in the apparent upregulation of RelE toxins (5).
Bioinformatic analysis revealed that M. tuberculosis H37Rv encodes 38 potential toxins (9 MazF, 3 RelE, 1 HigB, 2 ParE, and 23 VapC homologs), of which 34 are operonic with a potential antitoxin gene (44). In contrast, the related obligate intracellular parasite Mycobacterium leprae appears to have lost all functional toxin genes due to the unchanging nature of the niche M. leprae occupies in the human host (44). Zhu and coworkers reported that four of the MazF homologs (Rv2801c, Rv1991c, Rv1102c, and Rv0659c) caused cell growth arrest when overexpressed in E. coli and that two (Rv2801c and Rv1102c) had endoribonuclease activity both in vivo and in vitro (62, 63). Cleavage of single-stranded RNA by one of these MazF homologs from M. tuberculosis appears to have some sequence specificity, suggesting that it may play a direct role in posttranscriptional protein regulation (62). Rv1991c also has RNase activity (61), and overexpression of this protein is toxic in Mycobacterium smegmatis (7).
In this study, we sought to establish whether the fraction of drug-tolerant persisters in populations of M. tuberculosis cells represents a uniform population of metabolically inactive cells tolerant to multiple drugs (as has been proposed for E. coli persisters) (58). We investigated the role of the chromosomally encoded RelE toxins in the formation of drug-tolerant cells by both overexpression and targeted deletion. We show that the RelE proteins of M. tuberculosis are involved in determining the fraction of drug-tolerant persisters in vitro but that these persisters are drug and RelE isoform specific. None of the individual toxins was required for persistence in vivo, and treatment of mice infected with such mutants did not increase the frequency of in vivo persisters. These results suggest that individual classes of persisters have unique attributes and that even targeting individual mechanistic classes of persisters may not significantly impact the rate of sterilization by multidrug antituberculosis chemotherapy.
MATERIALS AND METHODS
Culture conditions, plasmids, and generation of relE1, relE2, or relE3 mutant strains.
The plasmids and primers used in this study are described in Tables 1 and 2. The E. coli XL2-Blue strain was used for cloning. M. tuberculosis was grown in Middlebrook 7H9 broth (Becton Dickinson, NJ) supplemented with ADC (0.81 mg/ml NaCl, 5 mg/ml bovine serum albumin fraction V [EMD Chemicals, NJ], 2 mg/ml d-glucose, 0.02% glycerol, and 0.05% Tween 80) or on Middlebrook 7H11 agar plates supplemented with OADC enrichment (ADC with 0.0006% [vol/vol] oleic acid). All drugs and antibiotics in this study were from Sigma-Aldrich (St. Louis, MO). Antibiotics were used at the following concentrations: ampicillin at 100 μg/ml, kanamycin at 50 μg/ml for E. coli and 10 μg/ml for mycobacteria, and hygromycin at 200 μg/ml for E. coli and 50 μg/ml for mycobacteria.
TABLE 1.
List of strains and plasmids used in this study
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| H37Rv | Laboratory strain (ATCC 27294) of M. tuberculosis H37Rv | |
| ΔrelE1 strain | relE1 mutant strain of M. tuberculosis H37Rv | This work |
| ΔrelE2 strain | relE2 mutant strain of M. tuberculosis H37Rv | This work |
| ΔrelE2 attB::relE2 strain | relE2::hyg complemented with relE2 at the attB site | This work |
| ΔrelE3 strain | relE3 mutant strain of M. tuberculosis H37Rv | This work |
| ΔmazF1 strain | mazF1 mutant strain of M. tuberculosis H37Rv | This work |
| ΔmazF5 strain | mazF5 mutant strain of M. tuberculosis H37Rv | This work |
| ΔmazF6 strain | mazF6 mutant strain of M. tuberculosis H37Rv | This work |
| Plasmids | ||
| p2NIL | Cloning vector, Kmr | 45 |
| pGOAL17 | Plasmid carrying sacB and lacZ genes as a PacI fragment, Apr | |
| p2NrelE1KO | p2NIL vector carrying upstream and downstream regions of relE1, the hyg cassette, and the PacI cassette from pGOAL17 | This work |
| p2NrelE2KO | p2NIL vector carrying upstream and downstream regions of relE2, the hyg cassette, and the PacI cassette from pGOAL17 | This work |
| p2NrelE3KO | p2NIL vector carrying upstream and downstream regions of relE3, the hyg cassette, and the PacI cassette from pGOAL17 | This work |
| pCR2.1 | Cloning vector, Kmr Apr | Invitrogen |
| pExp5-NT | T7-based expression vector | Invitrogen |
| pNT-RelE1 | pExp5-NT carrying relE1 | This work |
| pNT-RelBE1 | pExp5-NT carrying relB1 and relE1 | This work |
| pNT-RelE2 | pExp5-NT carrying relE2 | This work |
| pNT-RelBE2 | pExp5-NT carrying relB2 and relE2 | This work |
| pNT-RelE3 | pExp5-NT carrying relE3 | This work |
| pNT-RelBE3 | pExp5-NT carrying relB3 and relE3 | This work |
| pTE-mcs-1 | E. coli-Mycobacterium shuttle vector, Hygr, Atc inducible in M. tuberculosis | Kind gift from Sabine Ehrt; 11 |
| pTE-relE1 | pTE-mcs-1 carrying relE1 | This work |
| pTE-relE2 | pTE-mcs-1 carrying relE2 | This work |
| pTE-relE3 | pTE-mcs-1 carrying relE3 | This work |
| pTE-mazF1 | pTE-mcs-1 carrying mazF1 | This work |
| pTE-mazF2 | pTE-mcs-1 carrying mazF2 | This work |
| pTE-mazF3 | pTE-mcs-1 carrying mazF3 | This work |
| pTE-mazF4 | pTE-mcs-1 carrying mazF4 | This work |
| pTE-mazF5 | pTE-mcs-1 carrying mazF5 | This work |
| pTE-mazF6 | pTE-mcs-1 carrying mazF6 | This work |
| pTE-mazF7 | pTE-mcs-1 carrying mazF7 | This work |
TABLE 2.
List of oligonucleotides used in this study
| Oligonucleotide function and target gene(s) or region | Forward primer (5′-3′) | Reverse primer (5′-3′) | Probe |
|---|---|---|---|
| Construction of mutant strains | |||
| relE1 upstream | GGGAAGCTTTCTGCCCGGATCGAACGC | GGGGCGGCCGCCGCGCGGTGTAACGGTTGCG | |
| relE1 downstream | GGGGCGGCCGCGCCGTACCCCGGGTACCGG | GGTTAATTAAGCTGGCTCGCCGAACACC | |
| relE2 upstream | GGAAGCTTGGCGGGTCGGCTCGTAGCG | GGAGGCCTCGCTCAGTGGGGGCGTCGC | |
| relE2 downstream | GGAGGCCTCAACTCACCGACGGGCGCTC | GGTTAATTAACGCGGCGGACCGATCGACC | |
| relE3 upstream | GGGAAGCTTAGAACCCCGGCCGGTATCAG | GGGGATATCTCTCACTCCTCGCCGCCGGC | |
| relE3 downstream | GGGGATATCGATTTGGGGGCTGGTGGTATTC | GGGTTAATTAACGTGGATTTCACGCTCGCCG | |
| mazF1 upstream | GGAAGCTTCCGGCGAGGACGCCATCG | GGGAGGCCTCGACTCCGTCGCCGACGGTC | |
| mazF1 downstream | GGGAGGCCTCTCTCGCAGGTTCCGCGTCG | GGGTTAATTAACGATCCGTTCTGGGACCGAC | |
| mazF5 upstream | GGGAAGCTTCACGGCGATGGCGG | GGGAGGCCTCCCGATATCACGTCAGCAAAC | |
| mazF5 downstream | GGGAGGCCTGAGTCTCCAGCCGCCC | GGGTTAATTAAATCTCCTTAATGTGTTCTCGG | |
| mazF6 upstream | GGGAAGCTTTGGTGTGCGACGGGTCGTGG | GGGAGGCCTTAGGTCTACAGCACTGCACC | |
| mazF6 downstream | GGGAGGCCTCGTACGACCTCGCGGG | GGGTTAATTAAGCTATGAGCCGCTGTTCAC | |
| Construction of overexpression strains | |||
| relE1 | GGGCATATGAGCGACGACCATCCCTAC | GGGGCGGCCGCTTAACGTGGCCGGCACGGGa | |
| relB1 and relE1 | GGGCATATGGCTGTTGTCCCACTGGGCG | GGGGCGGCCGCTTAACGTGGCCGGCACGGG | |
| relE2 | GGGCATATGCCTTACACCGTGCGGTTC | GGGGCGGCCGCCTATCGGCGGTAGATGTCCGCGa | |
| relB2 and relE2 | GGGCATATGCGGATACTGCCGATTTCG | GGGGCGGCCGCCTATCGGCGGTAGATGTCCGCG | |
| relE3 | GGGCATATGGTGAGAAGCGTCAACTTCG | GGGGCGGCCGCTCAGTAGTGGTATCGGGCCa | |
| relB3 and relE3 | GGGCATATGAGCATCAGTGCGAGCGAG | GGGGCGGCCGCTCAGTAGTGGTATCGGGCC | |
| mazF1 | GGGCATATGGTGATGCGCCGCGGTGAG | GGGGCGGCCGCCTACGACCATAAGTCGAGATGC | |
| mazF2 | GGGCATATGCTGCGCGGTGAGATCTG | GGGGCGGCCGCTCAGCTCGGCAGGGGAGACG | |
| mazF3 | GGGCATATGGTGATTAGTCGTGCCG | GGGGCGGCCGCTCAAAGGTCCAGTACGCGAC | |
| mazF4 | GGGCATATGATGCGGCGCGGTGAATTGTG | GGGGCGGCCGCTCAACACCCCGTGCTCGCCC | |
| mazF5 | GGGCATATGACCGCACTTCCGGCGCG | GGGGCGGCCGCTCATCGAGAGCAATCGACGG | |
| mazF6 | GGGCATATGCGACCTATCCACATCGC | GGGGCGGCCGCCTATGCCACCACCCAATCG | |
| mazF7 | GGGCATATGAACGCGCCGTTGCGTG | GGGGCGGCCGCTCATGGCCACGGTAGCCCCA | |
| RT-PCR analysis | |||
| relB1 | CGCCGAAGTTGAGCTGACA | GATGACCGTGCCGGGTTA | ACGAGCGCATCACG |
| relE1 | AATGACCTTGAAGGCCTCCA | GATGGCGTAGACGACGCG | TCAGCCCGCCGCGGTGATT |
| relB2 | TGCGGATACTGCCGATTTC | CGCGTCGACGAACTCATTG | ACGATCAAGGGCAAGC |
| relE2 | CACGTTCAGCGCGCG | AGGATCACTACCGTTGTGTGCTC | CGCCTGCTGTACCGGATTGACGA |
| relB3 | CGAGGCAGCGCCTGTT | CGGCTGGTGATCGGTATTG | CCACTCATCGAACAGG |
| relE3 | TGCCTGGGAGGACTTCTTGT | ATCCGACGGGCCGTTT | CTGGCTGGCCGCTGATCGC |
| sigA | GACGAGGAGATCGCTGAACC | TCGTCTTCATCCCAGACGAAA | CCGAAAAGGACAAGGCCTCCGG |
This primer was also used for synthesis of cDNA from mycobacterial RNA, done using Superscript reverse transcriptase.
Mutant strains of M. tuberculosis lacking the toxin activity associated with the RelE1, RelE2, or RelE3 toxin were generated by homologous recombination. For making mutant strains, upstream and downstream flanking regions of the gene of interest were PCR amplified using gene-specific primers and cloned using the vector p2NIL (45). A PacI fragment from pGOAL17 (45) containing the lacZ and sacB genes was cloned into the PacI site to facilitate counterselection. Various mutant strains were generated using previously published protocols (45). The disruption of each gene was confirmed by PCR using gene-specific primers and Southern blotting with chromosomal DNA.
For overexpression analysis of E. coli, M. tuberculosis relE1, relE2, or relE3 was PCR amplified using gene-specific primers (Table 2) and cloned into pExp5-NT (Invitrogen, Carlsbad, CA) with an N-terminal His6 tag. For coexpression analysis of E. coli, genes encoding RelE1, RelE2, or RelE3 toxins were PCR amplified along with their cognate antitoxins by using gene-specific primers and subsequently cloned into the pExp5-NT vector. The expression of recombinant proteins was induced by the addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside) to E. coli at an optical density at 650 nm (OD650) of 0.4 at 30°C for 3 h. For toxicity assays with mycobacteria, M. tuberculosis relE1, relE2, or relE3 PCR fragments were amplified and cloned into an anhydrotetracycline (Atc)-inducible vector, pTE-mcs-1 (11). For overexpression of toxins in mycobacteria, cultures were grown to early log phase (OD650 of 0.1) and expression of toxins was induced by the addition of 50 ng/ml of Atc.
Activity of RelE toxins in E. coli and mycobacteria.
For toxicity assays with E. coli, the expression of toxins either alone or with their cognate antitoxins was induced by the addition of 1 mM IPTG. After induction for 3 h, 10-fold serial dilutions were made and 5 μl of each dilution was spotted on LB plates with and without IPTG. For overexpression of toxins in mycobacteria, cultures were grown to an OD650 of 0.1. Toxin expression was induced by the addition of 50 ng/ml Atc. At various time points, an aliquot was removed, 10-fold serial dilutions were made, and 100 μl was plated in duplicate on Middlebrook 7H11 plates for CFU analysis. Toxicity assays were performed independently at least three times.
RNA extraction.
Mycobacteria were grown to an OD650 of 0.2 to 0.3 and subsequently exposed to various drugs (at 10× MIC), 5 mM H2O2, or 5 mM S-nitrosoglutathione (GSNO) for 4 h. The concentrations of drugs used in the study were 4 μg/ml for Rif, 1 μg/ml for INH, 10 μg/ml for levofloxacin (Levo), and 10 μg/ml for gentamicin (Gm). For nutritional stress, mycobacteria were resuspended at an OD650 of 0.4 in Tris-buffered saline (pH 7.5)-0.05% Tween 80 (TBST) for 4 days. Total RNA was extracted from M. tuberculosis as previously described (6). Extracted RNA was DNase I treated using an Ambion DNA free kit (Applied Biosystems, Austin, TX). Complete DNA digestion was verified by subjecting an aliquot to 40 cycles of PCR using the appropriate primers (Table 2). M. tuberculosis RNA from mice infected for 4 weeks was extracted by homogenization of lungs in Trizol (Invitrogen, Carlsbad, CA) using a Pro300D homogenizer. Bacilli were harvested by centrifugation at 4,000 rpm for 10 min at 4°C, and RNA was subsequently extracted as previously described (6).
Quantitative real-time PCR (RT-PCR) analysis.
First-strand cDNA synthesis was carried out using Superscript III reverse transcriptase (Invitrogen, Carlsbad, CA). Real-time quantitative PCR was performed with a 7500 real-time PCR system (Applied Biosystems, Austin, TX) using a TaqMan PCR core reagent kit (Applied Biosystems, Austin, TX) according to the manufacturer's protocol. The sequences of primers used for first-strand cDNA synthesis and subsequent amplification are given in Table 2. A standard curve ranging from 0.15 pg to 1.5 ng genomic DNA was included in the PCR for quantification reference. The mRNA levels for relB1, relE1, relB2, relE2, relB3, and relE3 were quantified after normalization of mRNA levels to the expression level for the housekeeping gene sigA (6).
Microarray analysis.
Expression ratios from GenePix data files were calculated for each feature in each channel as the median number of pixels subtracted by the median number of background pixels for one color channel divided by the same for the second channel. If the feature pixel median did not exceed the background pixel median by three standard deviations, the ratio value was set to “missing” unless the median signal intensity in one channel was 5-fold above the median background number of pixels plus three standard deviations. Similarity between treatment or toxin overexpression groups was established using unsupervised class discovery by hierarchical clustering, with the Pearson correlation as the similarity metric (12). Genes which characterized the various treatment groups were identified using the shrunken centroids of gene expression method (55).
Persistence assays with RelE-overexpressing and relE mutant M. tuberculosis strains.
The drug concentrations used for M. tuberculosis persistence assays were as follows: 4 μg/ml Rif, 1 μg/ml INH, 10 μg/ml Levo, and 10 μg/ml Gm. For persistence studies of overexpression strains, the expression of toxins was induced by the addition of Atc to a final concentration of 50 ng/ml. RelE-overexpressing and relE mutant strains were diluted to an OD650 of 0.01 to 0.02 in 7H9 medium containing drugs at the concentrations indicated above and incubated at 37°C for 7 days. Cells were subsequently harvested by centrifugation at 3,000 × g for 10 min, washed twice with antibiotic-free media, and resuspended in 1 ml of 7H9 medium. For CFU analysis, duplicate 10-fold serial dilutions were made and aliquots were plated. Percent survival was calculated from the median number of CFU/ml in the culture after incubation with the drug for 7 days divided by the number of CFU/ml in the culture before the addition of drug.
Infection of murine macrophages.
The mouse macrophage-like J774A.1 cell line was cultured in Dulbecco's modified Eagle's medium (DMEM) (Invitrogen, Carlsbad, CA) containing 10% fetal calf serum (FCS). Cells were seeded in a 24-well plate for infection at a cell density of 5 × 105 per well. Cells were infected with either the wild type or a mutant strain at a multiplicity of infection (MOI) of 1:10 (macrophage to bacteria). After 6 h, cells were washed twice with 1× Hank's balanced salt solution and overlaid with 2 ml DMEM supplemented with FCS (10%) and amikacin (20 μg/ml). At various time points, the medium was aspirated and the macrophages were lysed with 1 ml of 0.05% sodium dodecyl sulfate (SDS) for 15 min at room temperature, followed by thorough pipetting to release intracellular bacteria. Duplicate 10-fold serial dilutions were prepared and aliquots plated in duplicate on 7H11 plates for colony counting.
Mouse infections.
Prior to infection, mycobacteria were passed through 5-μm filters and subcultured in fresh 7H9 medium to an OD650 of 0.45. Six- to 8-week-old C57BL/6 mice were infected with the wild-type, ΔrelE1, ΔrelE2, or ΔrelE3 H37Rv strain via the aerosol route, which results in the implantation of approximately 100 bacilli into the lungs (previously described in reference 6). Bacterial numbers were counted at 1, 14, 28, 56, and 102 days postinfection (four mice per time point) by independently homogenizing the lungs and spleen in 2 ml of 7H9 medium and plating aliquots of 10-fold serial dilutions on 7H11 plates in duplicate.
For drug treatment of infected mice, infected mice were orally gavaged 4 weeks after infection with 10 mg/kg of body weight of Rif daily for up to 5 weeks of treatment. At various time points, bacterial numbers in the lungs and spleens of Rif-treated mice (four mice per time point) were determined as described above.
Microarray data accession numbers.
The microarray data have been deposited in the Gene Expression Omnibus database at NCBI (GEO, http://www.ncbi.nlm.nih.gov/geo) under GEO Platform accession number GSE13998 and sample accession numbers GSM351644 to GSM351675.
RESULTS
relE transcription in M. tuberculosis is responsive to a variety of stress conditions.
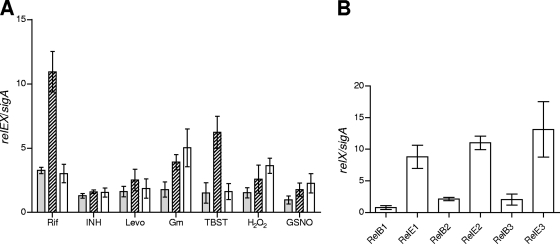
Three RelE homologs have been identified in the M. tuberculosis genome: Rv1246c (RelE1), Rv2866 (RelE2), and Rv3358 (RelE3) (44). These share 17.2%, 18.9%, and 15.8% amino acid identities, respectively, with E. coli RelE. M. tuberculosis RelE2 shares identities of 54% and 38% with M. tuberculosis RelE1 and RelE3, respectively. No significant identity was observed between the RelE1 and RelE3 proteins from M. tuberculosis. All M. tuberculosis RelE homologues appear to be cotranscribed with a short upstream open reading frame (ORF) which shares 21.3% (RelB1, Rv1247c), 21.5% (RelB2, Rv2865), and 19.8% (RelB3, Rv3357) identities with the E. coli RelB antitoxin. M. tuberculosis RelB1 shares 44% and 35% amino acid identities with the RelB2 and RelB3 proteins, respectively, whereas the RelB2 and RelB3 proteins share 30% identity. Since other RNA-cleaving toxins of the RelE family have been reported to be upregulated under various stress conditions, including nutrient deprivation and drug-induced stress (23, 27, 49), we first sought to understand whether M. tuberculosis relE toxins behaved similarly. We therefore directly determined the levels of mRNA transcripts under a variety of stress conditions by quantitative real-time PCR analysis (Fig. 1A). The amount of each of the three relE transcripts produced in the presence of various known antituberculosis drugs was normalized to that of the housekeeping gene sigA and compared to untreated-control ratios (6). We examined the responses to four different agents with differing modes of action: Rif, which inhibits transcription; Gm, which inhibits translation; Levo, which inhibits DNA gyrase; and the mycobacterium-specific drug INH, which affects primarily cell wall synthesis. The three genes of interest exhibited differential, overlapping specificities in their responses: relE1 was the most specific, and transcription was induced only 3-fold by Rif; relE2 mRNA levels were upregulated 11-, 4-, and 2.5-fold by Rif, Gm, and Levo, respectively; and relE3 was induced 3-, 5-, and 2-fold by Rif, Gm, and Levo, respectively (Fig. 1A). None of the three relE homologs were regulated in response to INH.
FIG. 1.
(A) Quantitative RT-PCR of relE1, relE2, and relE3 in response to various stresses. RT-PCR analysis was performed with total RNA extracted from in vitro-growing cultures, and expression levels were normalized to that for sigA. Data shown here are mean values and standard errors obtained from at least three independent experiments. relE1, filled bars; relE2, hatched bars; relE3, open bars. (B) Quantitative RT-PCR of relE toxins and relB antitoxins from lungs of infected mice. RT-PCR analysis was performed with total RNA extracted from mouse lungs, and expression levels were normalized to that for the housekeeping gene sigA. Data shown here are mean values and standard errors obtained from 3 different experiments.
Since M. tuberculosis encounters a number of adverse conditions, such as nutritional, oxidative, and nitrosative stress, in the host, the mRNA levels of relE1, relE2, and relE3 were evaluated under these conditions individually, as well as in bacteria isolated from the lungs of mice infected for 4 weeks. Similarly to results with drug-exposed cells, these stresses caused differential effects on the transcription of these relE-like genes, with nitrosative stress affecting relE3 transcription 2-fold, oxidative stress upregulating relE2 and relE3 levels 2.6-fold and 3.6-fold, respectively, and starvation in TBST resulting in the 6-fold upregulation of relE2 mRNA levels (Fig. 1A). Transcript levels of all three relE homologs, relE1, relE2, and relE3, were upregulated in lungs of mice infected for 4 weeks (9-, 11-, and 13-fold, respectively), whereas transcript levels of relB antitoxin homologs relB1, relB2, and relB3 were upregulated 0.8-, 2.0-, and 2.1-fold, respectively (Fig. 1B). This indicates that stresses other than nitrosative, oxidative, or nutritional stresses were responsible for the observed in vivo upregulation of relE1 transcription and that there may be added complexity to the regulation of these systems in the infected host.
Overexpression of the M. tuberculosis RelE homologs induces bacteriostasis.
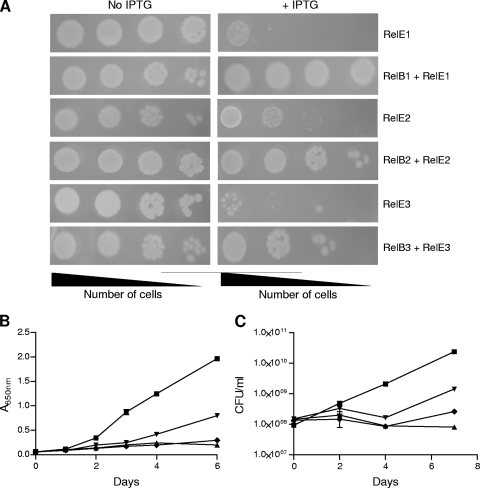
To explore the functional roles of the M. tuberculosis RelE homologs, all three were tested individually for toxicity upon overexpression in E. coli. Induction of protein expression by addition of 1 mM IPTG led to severe growth inhibition in cells overexpressing any of the three RelE homologs, whereas no growth inhibition was observed in cells containing the empty vector (Fig. 2A). This growth inhibition could partially be alleviated by coexpression of RelE1, RelE2, and RelE3 with their cognate RelB antitoxins (Fig. 2A). We also observed that overexpression of RelE2 and RelE3 induced a bacteriostatic effect on the growth of E. coli (data not shown).
FIG. 2.
The M. tuberculosis RelE homologs are metabolic toxins. (A) Toxicity assays with E. coli. E. coli BL21(λDE3)/plysS was transformed with plasmids expressing RelE1, RelE2, or RelE3 alone or coexpressed with their cognate antitoxins. Expression of toxins was induced with IPTG, with spotting of induced or noninduced cultures 3 h thereafter. (B and C) Growth inhibition by overexpression of RelE toxins in M. tuberculosis. Expression of RelE toxins in M. tuberculosis was induced by addition of Atc at a final concentration of 50 ng/ml. Viability of cells overexpressing RelE1 (triangles), RelE2 (inverted triangles), or RelE3 (diamonds) or vector-only controls (squares) was evaluated by measuring cell density at various OD650 (B) or CFU enumeration on 7H11 agar (C). The results shown are from one of three independent experiments.
To analyze the effect of RelE overexpression in M. tuberculosis, the ORFs were cloned under the control of an Atc-inducible promoter and transformed into the virulent H37Rv strain of M. tuberculosis (11). Consistently, RelE homolog expression inhibited cell growth (Fig. 2B). This was, however, a purely static effect, as no decrease in bacterial CFU was observed (Fig. 2C). A resumption of bacterial growth in cells overexpressing RelE2 and RelE3 was observed after 4 days (Fig. 2C), which may be due to reduced quantities of active Atc, since this regrowth could be prevented by further supplementation of cultures with Atc (data not shown). Bacterial CFU were reduced nearly 300-, 20-, and 100-fold in cells overexpressing RelE1, RelE2, and RelE3, respectively, compared to vector-only controls. Overexpression of M. tuberculosis RelE toxins in M. smegmatis achieved using Tet-inducible vectors severely inhibited bacterial growth, which could be reversed by coexpression of their cognate antitoxins (29).
The M. tuberculosis RelE toxins are individually nonessential for survival under in vitro and in vivo stress.
To evaluate the role of RelE1, RelE2, and RelE3 in survival under a variety of stresses, we constructed mutant strains lacking each individual ORF by gene replacement. For all of the mutant strains, the complete open reading frame was replaced by a hygromycin resistance gene, and the mutations were verified by Southern blotting (data not shown). All three mutant strains of M. tuberculosis had identical in vitro growth rates in Middlebrook 7H9 medium and were unimpaired in their survival during the long-term stationary phase and in their resistance to oxidative, nitrosative, or hypoxic stress conditions compared to the results for their wild-type parental counterpart strain (data not shown).
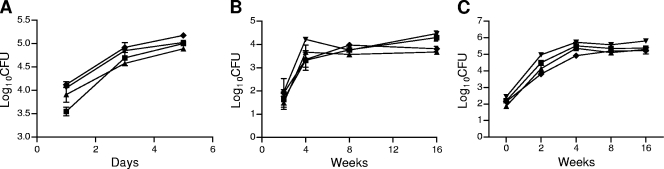
Surprisingly, in view of the observed upregulation of relE1, relE2, and relE3 in lung tissues of mice infected via the aerosol route, the mutant strains displayed growth rates and survival kinetics similar to those of the wild-type strain in J774A.1 macrophages (Fig. 3A). Similar results were also seen with the spleens and lungs of mice infected via the aerosol route (Fig. 3B and C). These data suggest that while these genes may play a functional role in the pathology of tuberculosis, they do not have a direct effect on the growth and survival of the bacterium itself.
FIG. 3.
RelE1, RelE2, and RelE3 are dispensable for in vivo survival of M. tuberculosis. (A) Growth of relE mutants in macrophages. J774A.1 resting macrophages were infected with the wild-type strain (H37Rv) (squares) or the ΔrelE1 (triangles), ΔrelE2 (inverted triangles), or ΔrelE3 (diamonds) mutant strain as described in Materials and Methods. At the designated time points, infected cells in triplicate wells were washed and lysed in phosphate-buffered saline (PBS) containing 0.05% SDS, and 100 μl of 10-fold serial dilutions was plated for CFU analysis. Results represent the means and standard errors of bacterial loads from three infected wells. (B and C) Growth and persistence of the relE mutants in mice. C57BL/6 mice were infected with the wild-type strain (H37Rv) (squares) or the ΔrelE1 (triangles), ΔrelE2 (inverted triangles), or ΔrelE3 (diamonds) mutant strain of M. tuberculosis. The bacterial loads were determined at the indicated time points postinfection by harvesting the spleens (B) and lungs (C) of infected mice. Results represent the medians and standard errors from four mice per group.
Transcriptional response to RelE overexpression.
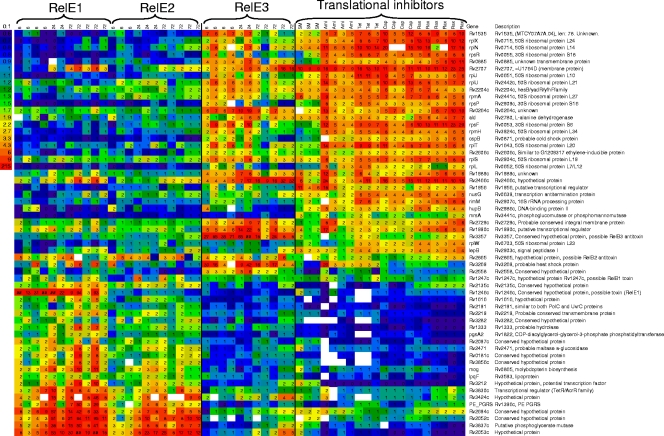
The drug-specific patterns in the transcriptional induction of the M. tuberculosis relE homologs led us to explore the transcriptional patterns of cells following intoxication by overexpression of each RelE protein. Transcriptional profiling of M. tuberculosis during overexpression of RelE3 indicated that the transcriptional response to RelE3 overexpression bore close similarities to that induced by translational inhibitors (Fig. 4). This response is characterized by the upregulation of several ribosomal and ribosome-associated proteins as well as genes encoding proteins that characterize the heat shock response (5). A subset of the genes that were elicited by RelE1 and RelE2 overexpression (i.e., the Rv2052c-Rv2053c and Rv2662-Rv2665 operons) have also been found to be upregulated under conditions of nutrient starvation (3, 5). However, in contrast to results with RelE3, the global profiles elicited by RelE1 and RelE2 overexpression were distinct from the transcriptional responses seen during treatment with the drugs that induced their expression (5) and did not elicit metabolic pathways that were suggestive of known physiologic conditions (5).
FIG. 4.
Transcriptional profiles of cells overexpressing toxins. Heat map-rendered table of expression changes for those genes that predictively associate overexpression of the RelE3 toxin with translational inhibitors or the RelE1 and RelE2 toxins. The inset shows the color scale for the expression ratios. Toxin overexpression was induced with Atc for 6, 24, or 72 h (as indicated above each column of expression ratios). The ratios represent levels of gene expression for Atc-induced strains compared to those for noninduced strains. Gene expression profiles obtained during translational inhibition have been reported previously (5). SM, streptomycin; Ami, amikacin; Tet, tetracycline; Cap, capreomycin; Rox, roxithromycin. Genes that were predictive for toxin overexpression compared to translational inhibition were identified by the nearest shrunken centroid method of Tibshirani et al. (55).
M. tuberculosis RNase toxins contribute to persister formation in vitro.
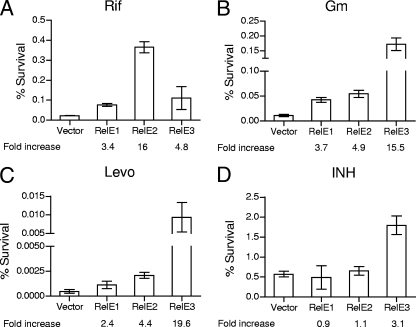
To determine the contribution of the M. tuberculosis RelE homologs to the formation of drug-tolerant persisters, RelE toxins were overexpressed by the addition of Atc for 4 days, and the fractions of persisters in M. tuberculosis overexpressing the toxins or in vector control cells were calculated after 1 week of exposure to the same four drugs used to test for induction of transcription (Rif, Gm, Levo, and INH) (Fig. 5). Overexpression of all three RelE homologs in M. tuberculosis increased persistence in the face of transcriptional and translational inhibition by Rif and Gm. RelE1 overexpression had the most modest effects overall on the fraction of persisters remaining after treatment with various drugs. RelE2 overexpression showed the greatest effect, increasing the number of Rif-tolerant persisters approximately 2-, 6- (P < 0.005), and 13 ± 3-fold (P < 0.0001) after 1, 2, and 4 days of toxin induction, respectively, consistent with our observation that relE2 was the most upregulated toxin upon Rif exposure (as determined by RT-PCR analysis). This effect was specific for RelE2 induction, since we did not see any difference in Rif-tolerant persisters in cells overexpressing RelE1 or RelE3 and in vector control cells during the course of the experiment (Fig. 5A; also see Fig. S2A in the supplemental material). RelE3 overexpression, in contrast, led to the greatest statistically significant change (15-fold) in persistence during Gm and Levo exposure (P < 0.001) (Fig. 5B and C). Overexpression of RelE3 also conferred a slight increase (P < 0.05) (Fig. 5D) in the frequency of persisters remaining after INH exposure. In contrast to the increase in numbers of persisters seen in stationary-phase cultures of E. coli, Staphylococcus aureus, and Pseudomonas aeruginosa (26), M. tuberculosis cultures did not have different fractions of Rif- and Levo-tolerant persisters in early log, log, and stationary phases (see Fig. S2B in the supplemental material), lending further support to the notion that the differences observed upon toxin overexpression were specific for the relE genes and not a function of the cell densities of the cultures.
FIG. 5.
Overexpression of the M. tuberculosis RelE homologs confers increased in vitro persistence to specific drugs. Mycobacteria harboring pTE-mcs-1, pTE-RelE1, pTE-RelE2, or pTE-RelE3 were grown to early log phase, and expression of toxins was induced by addition of Atc. After 4 days, the cultures were diluted to an OD650 of 0.02 to 0.03 in medium containing Rif (4 μg/ml), Gm (10 μg/ml), Levo (10 μg/ml), or INH (1 μg/ml). Persisters were harvested by centrifugation 7 days thereafter, washed, and plated for CFU analysis. Percent survival was measured as the number of CFU/ml in the culture after incubation with the drug relative to the number of CFU/ml in the culture before the addition of drug. Fold increase values are the ratios of RelE1, RelE2, or RelE3 survivors to “vector-only” survivors. The results shown are from one of three independent experiments.
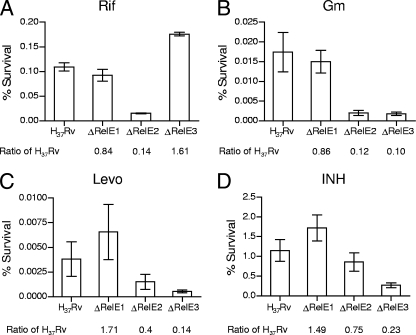
If overexpression of the RelE homologs increased persister formation and was responsible for generating persisters, then deletion of each gene individually should yield the opposite results. To complement and confirm the results from Fig. 5, we deleted each of the relE homologs individually to examine their effect on persister populations (Fig. 6). Despite modest increases in persister populations with RelE1 overexpression, the deletion of relE1 did not significantly affect persister populations after killing by any antibiotic agent. Deletion of relE2 significantly reduced the number of persisters 7-fold following treatment with Rif (Fig. 6A) (P < 0.001) and 8-fold following treatment with Gm (Fig. 6B) (P < 0.05), consistent with their increasing populations when RelE2 was overexpressed (Fig. 5A and B). Insignificant decreases were seen with Levo (Fig. 6C) and INH (Fig. 6D) treatment, although RelE2 overexpression increased survival rates only modestly (Fig. 5C and D). Conversely, relE3 deletion had marked effects on persister populations following killing with Gm (9-fold) (P < 0.05), Levo (4-fold) (P < 0.05), and INH (4-fold) (P < 0.05). The decrease in persister formation observed with INH in the relE3 deletion mutant was not seen with other cell wall-active agents, including ethambutol, thiolactamycin, cephradine-clavulanic acid, and pyrazinamide (data not shown).
FIG. 6.
Functional deletion of the M. tuberculosis RelE homologs confers decreased persistence to specific drugs. Early-log-phase cultures were diluted to an OD650 of 0.01 to 0.02 and treated with Rif (4 μg/ml), Gm (10 μg/ml), Levo (10 μg/ml), or INH (1 μg/ml) for 7 days at 37°C, followed by CFU analysis. CFU enumeration and percent survival were calculated as described in the legend for Fig. 5. The results shown are from one of three independent experiments.
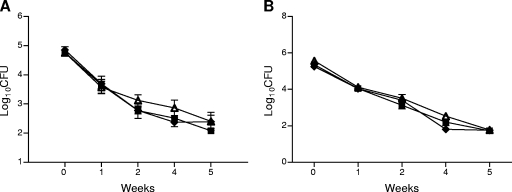
RelE2 does not contribute to rifampin persistence in vivo.
Since our results indicated that RelE2 plays a role in Rif persistence in vitro, we also investigated the physiological role of this toxin in persistence during Rif treatment of mice infected with the wild type, the ΔrelE2 strain, or the ΔrelE2 strain complemented with relE2. Infected mice were treated with 10 mg/kg of Rif daily for 5 weeks, and bacterial burdens in organs were quantified on a weekly basis (Fig. 7). These concentrations of Rif result in a peak blood serum concentration of 6.3 μg/ml at 1 h after administration (L. E. Via and C. E. Barry, unpublished results), which is well above the MIC. However, unlike the clear in vitro phenotype observed with the relE2 mutant, no persistence defect was observed in the lungs or spleens of infected mice treated with Rif.
FIG. 7.
Killing of wild-type, ΔrelE2, and ΔrelE2::relE2 strains in Rif-treated mice. C57BL/6 mice were infected with the wild-type strain (squares), the ΔrelE2 strain (triangles), or the complemented strain (diamonds) as described in Materials and Methods. After 4 weeks of infection, mice were orally gavaged with 10 mg/kg of Rif for 5 weeks. At the indicated time points, 4 mice per group were euthanized and bacterial loads in spleens (A) and lungs (B) of Rif-treated animals were determined.
Persisters to one drug are not cross-tolerant to other drugs.
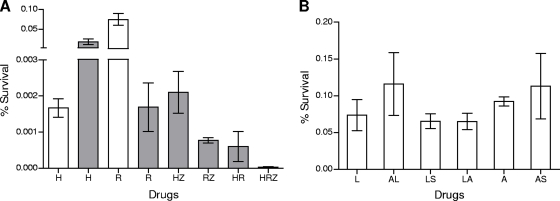
To determine whether the drug-tolerant M. tuberculosis persisters represented a uniformly multidrug-resistant population, early-log-phase cultures of M. tuberculosis (∼108 cells) were exposed to INH, Rif, and PZA at concentrations 10-fold higher than their MICs (Fig. 8A). In agreement with published literature, maximal INH-mediated killing was achieved at day 4 (23a). Shortly thereafter, INH-resistant mutants began to emerge due to the high mutation frequency associated with this drug, increasing the M. tuberculosis population more than 10-fold by day 7. Rif-mediated killing continued through day 7, with a final population representing 0.0016% of the input bacilli surviving. As expected, PZA killed less than 90% of the cells (results not shown).
FIG. 8.
(A) Preexisting drug-tolerant cells are not multidrug tolerant. The bactericidal activities of frontline drugs were evaluated against logarithmically growing cultures of M. tuberculosis either alone or in combination for either 4 days (open bars) or 7 days (filled bars). The drugs used in the experiment were isoniazid (H), rifampin (R), and pyrazinamide (Z) at 1, 4, and 200 μg/ml, respectively. Percent survival was measured as the number of CFU/ml in the culture after incubation with the drug relative to the number of CFU/ml in the culture pretreatment. The results shown are from one of three independent experiments. (B) Persisters in E. coli are multiply drug tolerant. Early-log-phase cultures of E. coli were exposed to a single drug for 3 h. After 3 h of exposure, a second drug was added for an additional 3 h. The drugs used in this experiment were levofloxacin (L), streptomycin (S), and ampicillin (A) at concentrations of 10 μg/ml, 24 μg/ml, and 100 μg/ml, respectively. Following drug exposures, the surviving bacilli were harvested by centrifugation and washed, and bacterial counts were determined by plating on agar medium.
The M. tuberculosis inocula were also exposed to various combinations of these three drugs (Fig. 8A). PZA did not reduce M. tuberculosis survival when used in concert with INH relative to the level seen with INH alone (compared to the maximal killing at day 4), while the combination of PZA and Rif did reduce survival compared to that found with Rif alone (day 7). Notably, the combination of INH and Rif reduced M. tuberculosis survival compared to the maximal killing seen with either INH or Rif alone. More strikingly, the combination of all three drugs resulted in a severe reduction in the survival of M. tuberculosis (2.8 × 10−5%) compared to that found with any single drug treatment. These results suggest that persisters surviving exposure to one antibiotic remain susceptible to other antibiotics. These results could be recapitulated by sequential addition of drugs to the fraction of persistent cells that survived 7-day exposure to the first drug (results not shown). In contrast, persister populations of E. coli were multidrug tolerant (Fig. 8B), as has been demonstrated earlier (58), indicating that E. coli persister populations are phenotypically more homogeneous.
DISCUSSION
Numerous studies have demonstrated that both slowly growing bacteria and bacteria surviving an initial drug challenge acquire phenotypic resistance to multiple antibiotics (2, 13, 26, 27, 32, 52, 58). Similarly, it has also been reported that nutrient-starved M. tuberculosis becomes resistant to multiple classes of drugs (3, 59). In vitro models for screening drugs against these persister populations have been based on, for example, bacterial populations surviving Rif killing (40). This study was an initial attempt to address the hypothesis that drug-tolerant persisters were a uniform subpopulation of M. tuberculosis cells under the influence of one of the multitude of toxin-antitoxin modules present in M. tuberculosis. The previous association of the RelE family RNase toxins with nutrient starvation and persister formation in other organisms (27), combined with our observation that all three relE homologs were induced by multiple drug classes in vitro (5), led us to study this family in detail.
We refined our initial study of the transcriptional regulation of the relE family genes in M. tuberculosis by using quantitative RT-PCR and found that expression of each of the three relE toxins was responsive to specific stress conditions. The breadth and specificity of the regulation were surprising. Although relE1 responded to inhibition of transcription (by Rif) in vitro, the highest upregulation was displayed by relE2, which was upregulated 11-fold during transcriptional inhibition (although it did also respond to other stresses). In contrast, relE3 was the most responsive to translational inhibition, as seen by its 5-fold upregulation by Gm, although it was responsive to a lesser extent to other stresses. Interestingly, all three toxins, RelE1, RelE2, and RelE3, were highly upregulated between 9- and 13-fold in the lungs of mice that had been infected for 4 weeks, whereas the corresponding three antitoxins, RelB1, RelB2, and RelB3, demonstrated considerably less upregulation (0.8- to 2-fold) in vivo. This implies that the level of antitoxin produced during host pathogenesis is not sufficient to neutralize its cognate toxin and that, as a result, macromolecular metabolism is suppressed. It is likely that the regulation of the toxins in vivo depends on the local environment of the bacterium, with the transcriptional analyses representing only an average of the heterogeneous populations. This implies that the signals to which the different toxins respond have subtle, but important, differences.
As expected, heterologous overexpression of RelE1, RelE2, and RelE3 in E. coli strongly inhibited the growth of this organism, and the toxicity of these proteins was reduced when they were coexpressed with their cognate antitoxin partners. These toxins had similar activities when their expression was induced in M. tuberculosis. However, unlike reports describing a bactericidal activity induced by overexpression of some MazF toxins (1, 14, 16, 28, 42), overexpression of the M. tuberculosis RelE toxins led to a state of reversible bacteriostasis. Similarly, we have found that inducible overexpression of the MazF homologs Rv2801c (MazF1), Rv1991c (MazF3), and Rv1102c (MazF6) in M. tuberculosis was bacteriostatic (see Fig. S1A in the supplemental material), suggesting that RelE and MazF toxins do not function to promote cell death in M. tuberculosis. We cannot exclude, of course, the possibility that these toxins may result in cell death by acting in concert with another protein or under specific environmental conditions (see Fig. S1A in the supplemental material).
Ectopic overexpression and gene deletion of each of these toxins led to consistent, specific changes in the fraction of persister cells remaining following killing by individual antituberculosis drugs. Each of the different toxins contributed to the generation of persister cells with markedly different phenotypes. RelE2 overexpression, for example, led to an increase in Rif-tolerant cells, and deletion of this toxin led to a decrease in such persisters. Persisters to INH exposure were affected by RelE3, with overexpression and deletion of this toxin resulting in increased and decreased numbers, respectively. Both RelE2 and RelE3 affected persister cell formation following Levo and Gm exposure. RelE1 appeared to be the exception, with overexpression increasing the fraction of persisters only modestly during a variety of drug exposures and deletion failing to show an effect on persistence to any agent (Fig. 5 and 6 and data not shown). Although the level of persistence observed in M. tuberculosis overexpressing the RelE toxins did not achieve the same levels as that reported for E. coli overexpressing RelE (27), the copy number of the mycobacterial plasmids based on the oriM origin of replication is 3 to 5 (47), which is lower than the copy number of 12 to 15 for the plasmids used for RelE expression in E. coli; different promoters were used to drive expression in these systems. In contrast, deletion of relE in E. coli did not lead to a persistence defect (27), whereas deletion of the mycobacterial homolog led to drug-specific persistence defects in vitro. These results demonstrate that while persistence can be induced by overexpression of these RelE-like toxins, the metabolic state induced by any single toxin is clearly not one of generalized metabolic shutdown. Likewise, we have found that overexpression and knockout of several of the M. tuberculosis mazF genes, Rv1102c (mazF6), Rv1942c (mazF5), and Rv2801c (mazF1), gave substantially similar results in that deletion or overexpression gave rise to drug-specific effects on persister formation (data not shown).
The difference in response among the three M. tuberculosis RelE homologs most likely suggests that they have different mRNA substrates. This is perhaps not surprising given the specificity of other endoribonuclease systems, such as the kid and kis system of the parD operon of E. coli, as well as the sequence specificity of MazF (25, 60). Indeed, at least two of the MazF homologs of M. tuberculosis have been shown to have slightly different RNA consensus cleavage sites (62). This result provides at least a conceptual framework for rationalizing the differential effects of these toxins on the global phenotype of bacterial cells, as each toxin may have a separate, defined subset of the transcriptome with which it interacts. The relative abundance of toxin-antitoxin loci in M. tuberculosis may therefore reflect the need for an additional level of control over the expression of specific protein subsets in comparison to that for organisms such as E. coli, which has only 5 modules (44), in addition to general mechanisms for the regulation of transcription in this slowly growing pathogen. The differing antibiotic susceptibilities that result may simply be a downstream consequence of alterations in protein levels of either target proteins or activators.
An analysis of the transcriptional response to upregulation of these toxins under the control of the tetracycline promoter provides a glimpse of the normal transcript ensemble they degrade. RelE3 clustered closely with inhibitors of translation, as evidenced by the large-scale upregulation of genes associated with translation, suggesting that this toxin may be involved in switching off protein synthesis. Intriguingly, RelE3 was associated with the strongest persistence phenotype with translational inhibitors. The translational response of the cells as a result of RelE3 overexpression was not surprising since the RelE toxin of E. coli has been shown to cleave mRNA in the context of the ribosome. The transcriptional response elicited by upregulation of the RelE1 and RelE2 homologs did not cluster with any known inhibitors of metabolism, although they had genes in common with those upregulated upon nutrient starvation in M. tuberculosis (3). Complete nutrient starvation in vitro is, of course, an extreme case, and the organism may well have evolved differential responses to the depletion of different nutrients. Interestingly, the transcriptional profile of cells overexpressing RelE1 differed from that of cells overexpressing RelE2 by its downregulation of genes encoding the ATP synthase, suggesting differences in energy metabolism between these cells. In all three cases, we observed upregulation of the cognate antitoxin to each homolog of RelE, most likely reflecting an inherent homeostatic property of this regulatory mechanism.
Although all three toxins were upregulated during in vivo growth within mouse lungs, all three toxins individually were dispensable for replication and chronic persistence in mouse tissues. Similarly, we found that knockout mutants of the mazF genes encoded by Rv1102c (mazF6), Rv2801c (mazF1), and Rv1942c (mazF5) were not compromised for growth or persistence in lungs of infected mice (see Fig. S1B in the supplemental material). These paradoxical results suggest either that there is redundancy in the control of metabolism by cross talk between the homologs (or other regulatory systems) or that the fitness defect of a failure to adjust transcript levels is subtle. The mouse model, of course, represents only a small fraction of the physiological diversity of conditions to which M. tuberculosis is normally exposed, so such a phenotype may not be apparent. Even the apparent defect in persister formation observed in vitro did not translate in vivo, since treatment of relE2-infected mice with 10 mg/kg of Rif did not result in any difference in kinetics of bacterial killing or final bacterial burden during 5 weeks of treatment. Thus, our results show that there are fundamental differences between drug-tolerant persisters of E. coli and M. tuberculosis. In E. coli, drug persisters that survive exposure to bactericidal concentrations of one antibiotic are broadly tolerant to antibiotics of different classes (58), as recapitulated in this study (Fig. 8B), whereas in M. tuberculosis cultures, preexisting drug-tolerant persisters represent different populations of cells that remain sensitive to other drug classes (Fig. 8A).
The implications of these findings are clear from such simple experiments using the three mainstays of antituberculosis chemotherapy that are coadministered to the majority of tuberculosis patients globally. Our in vitro results suggest that persisters to any one drug are not automatically persisters to another drug. The fact that individual toxins are associated with drug-specific persistence phenotypes further supports the notion that persisters can arise by a multitude of mechanisms. Whether any subset of persister cells plays a significant role in determining the duration of antituberculosis chemotherapy depends on the spectrum of mechanisms that are engaged in those cells. It remains possible, given the complexities of differential drug penetration into the various types of lesions found in humans, that a subset of the potential persister phenotypes is relevant to chemotherapy duration, but identifying whether and which of the 38 potential RNase toxins play a role in generating the persisters responsible for the relapse of disease in humans will be experimentally challenging.
Supplementary Material
Acknowledgments
Funding for this work was provided by the Division of Intramural Research, NIAID, NIH.
We also acknowledge the technical help of Jacqueline Gonzales in performing animal experiments and in vitro persistence studies.
Footnotes
Published ahead of print on 8 January 2010.
Supplemental material for this article may be found at http://jb.asm.org/.
REFERENCES
- 1.Amitai, S., Y. Yassin, and H. Engelberg-Kulka. 2004. MazF-mediated cell death in Escherichia coli: a point of no return. J. Bacteriol. 186:8295-8300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Balaban, N. Q., J. Merrin, R. Chait, L. Kowalik, and S. Leibler. 2004. Bacterial persistence as a phenotypic switch. Science 305:1622-1625. [DOI] [PubMed] [Google Scholar]
- 3.Betts, J. C., P. T. Lukey, L. C. Robb, R. A. McAdam, and K. Duncan. 2002. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 43:717-731. [DOI] [PubMed] [Google Scholar]
- 4.Boshoff, H. I., and C. E. Barry III. 2005. Tuberculosis—metabolism and respiration in the absence of growth. Nat. Rev. Microbiol. 3:70-80. [DOI] [PubMed] [Google Scholar]
- 5.Boshoff, H. I., T. G. Myers, B. R. Copp, M. R. McNeil, M. A. Wilson, and C. E. Barry III. 2004. The transcriptional responses of Mycobacterium tuberculosis to inhibitors of metabolism: novel insights into drug mechanisms of action. J. Biol. Chem. 279:40174-40184. [DOI] [PubMed] [Google Scholar]
- 6.Boshoff, H. I., M. B. Reed, C. E. Barry III, and V. Mizrahi. 2003. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell 113:183-193. [DOI] [PubMed] [Google Scholar]
- 7.Carroll, P., A. C. Brown, A. R. Hartridge, and T. Parish. 2007. Expression of Mycobacterium tuberculosis Rv1991c using an arabinose-inducible promoter demonstrates its role as a toxin. FEMS Microbiol. Lett. 274:73-82. [DOI] [PubMed] [Google Scholar]
- 8.Christensen, S. K., M. Mikkelsen, K. Pedersen, and K. Gerdes. 2001. RelE, a global inhibitor of translation, is activated during nutritional stress. Proc. Natl. Acad. Sci. U. S. A. 98:14328-14333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christensen, S. K., K. Pedersen, F. G. Hansen, and K. Gerdes. 2003. Toxin-antitoxin loci as stress-response-elements: ChpAK/MazF and ChpBK cleave translated RNAs and are counteracted by tmRNA. J. Mol. Biol. 332:809-819. [DOI] [PubMed] [Google Scholar]
- 10.Dahl, J. L., C. N. Kraus, H. I. Boshoff, B. Doan, K. Foley, D. Avarbock, G. Kaplan, V. Mizrahi, H. Rubin, and C. E. Barry III. 2003. The role of RelMtb-mediated adaptation to stationary phase in long-term persistence of Mycobacterium tuberculosis in mice. Proc. Natl. Acad. Sci. U. S. A. 100:10026-10031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ehrt, S., X. V. Guo, C. M. Hickey, M. Ryou, M. Monteleone, L. W. Riley, and D. Schnappinger. 2005. Controlling gene expression in mycobacteria with anhydrotetracycline and Tet repressor. Nucleic Acids Res. 33:e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisen, M. B., P. T. Spellman, P. O. Brown, and D. Botstein. 1998. Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U. S. A. 95:14863-14868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Eng. R. H., F. T. Padberg, S. M. Smith, E. N. Tan, and C. E. Cherubin. 1991. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 35:1824-1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Engelberg-Kulka, H., R. Hazan, and S. Amitai. 2005. mazEF: a chromosomal toxin-antitoxin module that triggers programmed cell death in bacteria. J. Cell Sci. 118:4327-4332. [DOI] [PubMed] [Google Scholar]
- 15.Evans, D. J., M. R. Brown, D. G. Allison, and P. Gilbert. 1990. Susceptibility of bacterial biofilms to tobramycin: role of specific growth rate and phase in the division cycle. J. Antimicrob. Chemother. 25:585-591. [DOI] [PubMed] [Google Scholar]
- 16.Fu, Z., N. P. Donegan, G. Memmi, and A. L. Cheung. 2007. Characterization of MazFSa, an endoribonuclease from Staphylococcus aureus. J. Bacteriol. 189:8871-8879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garton, N. J., S. J. Waddell, A. L. Sherratt, S. M. Lee, R. J. Smith, C. Senner, J. Hinds, K. Rajakumar, R. A. Adegbola, G. S. Besra, P. D. Butcher, and M. R. Barer. 2008. Cytological and transcript analyses reveal fat and lazy persister-like bacilli in tuberculous sputum. PLoS Med. 5:e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gerdes, K., S. K. Christensen, and A. Lobner-Olesen. 2005. Prokaryotic toxin-antitoxin stress response loci. Nat. Rev. Microbiol. 3:371-382. [DOI] [PubMed] [Google Scholar]
- 19.Gomez, J. E., and J. D. McKinney. 2004. M. tuberculosis persistence, latency, and drug tolerance. Tuberculosis (Edinb.) 84:29-44. [DOI] [PubMed] [Google Scholar]
- 20.Gotfredsen, M., and K. Gerdes. 1998. The Escherichia coli relBE genes belong to a new toxin-antitoxin gene family. Mol. Microbiol. 29:1065-1076. [DOI] [PubMed] [Google Scholar]
- 21.Grosset, J., C. Truffot-Pernot, C. Lacroix, and B. Ji. 1992. Antagonism between isoniazid and the combination pyrazinamide-rifampin against tuberculosis infection in mice. Antimicrob. Agents Chemother. 36:548-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hayes, F. 2003. Toxins-antitoxins: plasmid maintenance, programmed cell death, and cell cycle arrest. Science 301:1496-1499. [DOI] [PubMed] [Google Scholar]
- 23.Hazan, R., B. Sat, and H. Engelberg-Kulka. 2004. Escherichia coli mazEF-mediated cell death is triggered by various stressful conditions. J. Bacteriol. 186:3663-3669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23a.Herbert, D., C. N. Paramasivan, P. Venkatesan, G. Kubendiran, R. Prabhakar, and D. A. Mitchison. 1996. Bactericidal action of ofloxacin, sulbactam-ampicillin, rifampin, and isoniazid on logarithmic- and stationary-phase cultures of Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 40:2296-2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ivanova, N., M. Y. Pavlov, B. Felden, and M. Ehrenberg. 2004. Ribosome rescue by tmRNA requires truncated mRNAs. J. Mol. Biol. 338:33-41. [DOI] [PubMed] [Google Scholar]
- 25.Kamphuis, M. B., M. C. Monti, R. H. van den Heuvel, S. Santos-Sierra, G. E. Folkers, M. Lemonnier, R. Diaz-Orejas, A. J. Heck, and R. Boelens. 2007. Interactions between the toxin Kid of the bacterial parD system and the antitoxins Kis and MazE. Proteins 67:219-231. [DOI] [PubMed] [Google Scholar]
- 26.Keren, I., N. Kaldalu, A. Spoering, Y. Wang, and K. Lewis. 2004. Persister cells and tolerance to antimicrobials. FEMS Microbiol. Lett. 230:13-18. [DOI] [PubMed] [Google Scholar]
- 27.Keren, I., D. Shah, A. Spoering, N. Kaldalu, and K. Lewis. 2004. Specialized persister cells and the mechanism of multidrug tolerance in Escherichia coli. J. Bacteriol. 186:8172-8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kolodkin-Gal, I., and H. Engelberg-Kulka. 2006. Induction of Escherichia coli chromosomal mazEF by stressful conditions causes an irreversible loss of viability. J. Bacteriol. 188:3420-3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Korch, S. B., H. Contreras, and J. E. Clark-Curtiss. 2009. Three Mycobacterium tuberculosis Rel toxin-antitoxin modules inhibit mycobacterial growth and are expressed in infected human macrophages. J. Bacteriol. 191:1618-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korch, S. B., T. A. Henderson, and T. M. Hill. 2003. Characterization of the hipA7 allele of Escherichia coli and evidence that high persistence is governed by (p)ppGpp synthesis. Mol. Microbiol. 50:1199-1213. [DOI] [PubMed] [Google Scholar]
- 31.Korch, S. B., and T. M. Hill. 2006. Ectopic overexpression of wild-type and mutant hipA genes in Escherichia coli: effects on macromolecular synthesis and persister formation. J. Bacteriol. 188:3826-3836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kussell, E., R. Kishony, N. Q. Balaban, and S. Leibler. 2005. Bacterial persistence: a model of survival in changing environments. Genetics 169:1807-1814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lewis, K. 2008. Multidrug tolerance of biofilms and persister cells. Curr. Top. Microbiol. Immunol. 322:107-131. [DOI] [PubMed] [Google Scholar]
- 34.Li, X., M. Yagi, T. Morita, and H. Aiba. 2008. Cleavage of mRNAs and role of tmRNA system under amino acid starvation in Escherichia coli. Mol. Microbiol. 68:462-473. [DOI] [PubMed] [Google Scholar]
- 35.Li, Y., and Y. Zhang. 2007. PhoU is a persistence switch involved in persister formation and tolerance to multiple antibiotics and stresses in Escherichia coli. Antimicrob. Agents Chemother. 51:2092-2099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.McCune, R. M., F. M. Feldmann, H. P. Lambert, and W. McDermott. 1966. Microbial persistence. I. The capacity of tubercle bacilli to survive sterilization in mouse tissues. J. Exp. Med. 123:445-468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCune, R. M., F. M. Feldmann, and W. McDermott. 1966. Microbial persistence. II. Characteristics of the sterile state of tubercle bacilli. J. Exp. Med. 123:469-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.McCune, R. M., Jr., W. McDermott, and R. Tompsett. 1956. The fate of Mycobacterium tuberculosis in mouse tissues as determined by the microbial enumeration technique. II. The conversion of tuberculous infection to the latent state by the administration of pyrazinamide and a companion drug. J. Exp. Med. 104:763-802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchison, D. A. 2000. Role of individual drugs in the chemotherapy of tuberculosis. Int. J. Tuberc. Lung Dis. 4:796-806. [PubMed] [Google Scholar]
- 40.Mitchison, D. A., and A. R. Coates. 2004. Predictive in vitro models of the sterilizing activity of anti-tuberculosis drugs. Curr. Pharm. Des. 10:3285-3295. [DOI] [PubMed] [Google Scholar]
- 41.Moyed, H. S., and K. P. Bertrand. 1983. hipA, a newly recognized gene of Escherichia coli K-12 that affects frequency of persistence after inhibition of murein synthesis. J. Bacteriol. 155:768-775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nariya, H., and M. Inouye. 2008. MazF, an mRNA interferase, mediates programmed cell death during multicellular Myxococcus development. Cell 132:55-66. [DOI] [PubMed] [Google Scholar]
- 43.Neff, M. 2003. ATS, CDC, and IDSA update recommendations on the treatment of tuberculosis. Am. Fam. Physician 68:1854-1862. [PubMed] [Google Scholar]
- 44.Pandey, D. P., and K. Gerdes. 2005. Toxin-antitoxin loci are highly abundant in free-living but lost from host-associated prokaryotes. Nucleic Acids Res. 33:966-976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parish, T., and N. G. Stoker. 2000. Use of a flexible cassette method to generate a double unmarked Mycobacterium tuberculosis tlyA plcABC mutant by gene replacement. Microbiology 146:1969-1975. [DOI] [PubMed] [Google Scholar]
- 46.Pedersen, K., A. V. Zavialov, M. Y. Pavlov, J. Elf, K. Gerdes, and M. Ehrenberg. 2003. The bacterial toxin RelE displays codon-specific cleavage of mRNAs in the ribosomal A site. Cell 112:131-140. [DOI] [PubMed] [Google Scholar]
- 47.Ranes, M. G., J. Rauzier, M. Lagranderie, M. Gheorghiu, and B. Gicquel. 1990. Functional analysis of pAL5000, a plasmid from Mycobacterium fortuitum: construction of a “mini” mycobacterium-Escherichia coli shuttle vector. J. Bacteriol. 172:2793-2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sahbazian, B., and S. E. Weis. 2005. Treatment of active tuberculosis: challenges and prospects. Clin. Chest Med. 26:273-282. [DOI] [PubMed] [Google Scholar]
- 49.Sat, B., M. Reches, and H. Engelberg-Kulka. 2003. The Escherichia coli mazEF suicide module mediates thymineless death. J. Bacteriol. 185:1803-1807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Scanga, C. A., V. P. Mohan, H. Joseph, K. Yu, J. Chan, and J. L. Flynn. 1999. Reactivation of latent tuberculosis: variations on the Cornell murine model. Infect. Immun. 67:4531-4538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schnappinger, D., S. Ehrt, M. I. Voskuil, Y. Liu, J. A. Mangan, I. M. Monahan, G. Dolganov, B. Efron, P. D. Butcher, C. Nathan, and G. K. Schoolnik. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shah, D., Z. Zhang, A. Khodursky, N. Kaldalu, K. Kurg, and K. Lewis. 2006. Persisters: a distinct physiological state of E. coli. BMC Microbiol. 6:53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Smith, P. A., and F. E. Romesberg. 2007. Combating bacteria and drug resistance by inhibiting mechanisms of persistence and adaptation. Nat. Chem. Biol. 3:549-556. [DOI] [PubMed] [Google Scholar]
- 54.Spoering, A. L., M. Vulic, and K. Lewis. 2006. GlpD and PlsB participate in persister cell formation in Escherichia coli. J. Bacteriol. 188:5136-5144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tibshirani, R., T. Hastie, B. Narasimhan, and G. Chu. 2002. Diagnosis of multiple cancer types by shrunken centroids of gene expression. Proc. Natl. Acad. Sci. U. S. A. 99:6567-6572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Via, L. E., P. L. Lin, S. M. Ray, J. Carrillo, S. S. Allen, S. Y. Eum, K. Taylor, E. Klein, U. Manjunatha, J. Gonzales, E. G. Lee, S. K. Park, J. A. Raleigh, S. N. Cho, D. N. McMurray, J. L. Flynn, and C. E. Barry III. 2008. Tuberculous granulomas are hypoxic in guinea pigs, rabbits, and nonhuman primates. Infect. Immun. 76:2333-2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Warner, D. F., and V. Mizrahi. 2006. Tuberculosis chemotherapy: the influence of bacillary stress and damage response pathways on drug efficacy. Clin. Microbiol. Rev. 19:558-570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wiuff, C., R. M. Zappala, R. R. Regoes, K. N. Garner, F. Baquero, and B. R. Levin. 2005. Phenotypic tolerance: antibiotic enrichment of noninherited resistance in bacterial populations. Antimicrob. Agents Chemother. 49:1483-1494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Xie, Z., N. Siddiqi, and E. J. Rubin. 2005. Differential antibiotic susceptibilities of starved Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 49:4778-4780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang, J., Y. Zhang, and M. Inouye. 2003. Characterization of the interactions within the mazEF addiction module of Escherichia coli. J. Biol. Chem. 278:32300-32306. [DOI] [PubMed] [Google Scholar]
- 61.Zhao, L., and J. Zhang. 2008. Biochemical characterization of a chromosomal toxin-antitoxin system in Mycobacterium tuberculosis. FEBS Lett. 582:710-714. [DOI] [PubMed] [Google Scholar]
- 62.Zhu, L., S. Phadtare, H. Nariya, M. Ouyang, R. N. Husson, and M. Inouye. 2008. The mRNA interferases, MazF-mt3 and MazF-mt7 from Mycobacterium tuberculosis target unique pentad sequences in single-stranded RNA. Mol. Microbiol. 69:559-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhu, L., Y. Zhang, J. S. Teh, J. Zhang, N. Connell, H. Rubin, and M. Inouye. 2006. Characterization of mRNA interferases from Mycobacterium tuberculosis. J. Biol. Chem. 281:18638-18643. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.