FIG. 1.
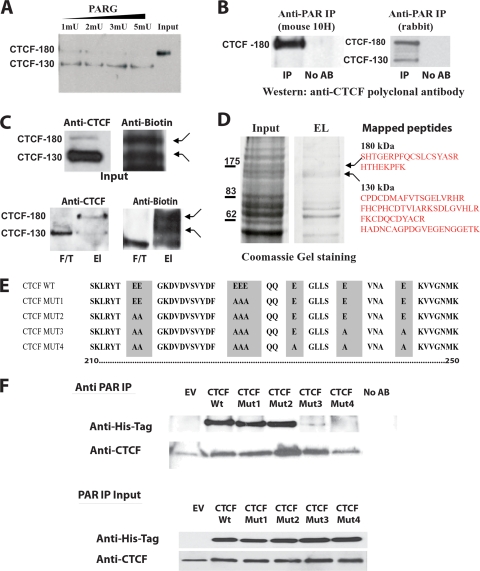
CTCF-180 and CTCF-130 are differentially poly(ADP-ribosyl)ated. (A) Removal of PARylation from CTCF-180 using PARG. MCF7 cell nucleoplasmic extracts containing only CTCF-180 were incubated with PARG enzyme in a dose-dependent manner. The incubated lysates were analyzed by Western blotting using an anti-CTCF polyclonal antibody (Abcam). A complete shift from CTCF-180 to CTCF-130 is observed following increases in units of PARG. (B) The anti-PAR rabbit polyclonal antibody and mouse 10H antibody immunoprecipitate specific forms of CTCF. MCF7 cell extracts were immunoprecipitated with either the anti-PAR rabbit polyclonal antibody or anti-PAR mouse (10H) antibody. The immunoprecipitated proteins were analyzed by Western blotting with the anti-CTCF polyclonal antibody. (Left) The anti-PAR mouse 10H antibody, which recognizes more than 20 PAR residues, immunoprecipitates only CTCF-180. (Right) The anti-PAR rabbit polyclonal antibody is able to recognize as few as two PAR residues and is able to immunoprecipitate both CTCF-130 and CTCF-180. No AB, control with no antibody in the IP reaction mixture. (C) Labeling with biotinylated NAD+ reveals two forms, CTCF-180 and CTCF-130. MCF7 cells (7.5 × 105) were incubated with biotinylated NAD+, leading to the incorporation of biotinylated ADP-ribose residues attached to protein acceptors. The labeled proteins were selected using M-280 streptavidin magnetic beads. (Top) The input was analyzed for the presence of the CTCF forms by a Western blot assay. The membrane was probed with the anti-CTCF rabbit polyclonal antibody (Abcam), stripped, and probed with antibiotin antibody. Both CTCF-130 and CTCF-180 forms were present in the extracts (left), and bands of similar sizes (indicated by arrows) were observed when the membrane was probed with the antibiotin antibody (right). (Bottom) For selected biotinylated proteins, the elution (El) and flowthrough (F/T) fractions were analyzed by a Western blot assay for the presence of CTCF. (Left) CTCF-130 appears in both fractions, whereas CTCF-180 is observed only in the elution fraction. (Right) The membrane was stripped and reprobed with the antibiotin antibody to confirm the presence of similarly sized labeled bands (indicated by arrows). (D) Analysis of the 130- and 180-kDa bands of the PARylated proteins selected using M-280 streptavidin magnetic beads and mass spectrometry. Proteins eluted from streptavidin magnetic beads were subjected to SDS-PAGE and stained with Coomassie blue. Bands corresponding to 130 and 180 kDa were excised and analyzed by mass spectrometry. The identified CTCF peptides are shown. (E) Schematic diagram of the CTCF N-terminal domain amino acid sequence showing the mutations produced by site-directed mutagenesis. The wild-type sequence is depicted at the top. Amino acid substitutions are highlighted. The amino acids are numbered according to Filippova et al. (32). (F) Western blot analysis of the proteins produced by the mutant variants of CTCF. 293T cells were transfected with 5 μg of EV or a vector expressing the CTCF WT or a CTCF mutant. Twenty-four hours posttransfection, cells were lysed and the cell extracts were divided in two and either subjected to IP with the anti-PAR rabbit polyclonal antibody or used as an input control. (Top) The immunoprecipitated proteins were resolved by SDS-PAGE, blotted, and probed with the anti-His tag antibody. The membrane was reprobed with an anti-CTCF antibody, confirming the presence of endogenous CTCF-130, retained by the anti-PAR antibody, in all cell lysates. (Bottom) The input materials were resolved by SDS-PAGE and probed with both the anti-His tag antibody and anti-CTCF, showing equal amounts of the exogenous and endogenous CTCF proteins.