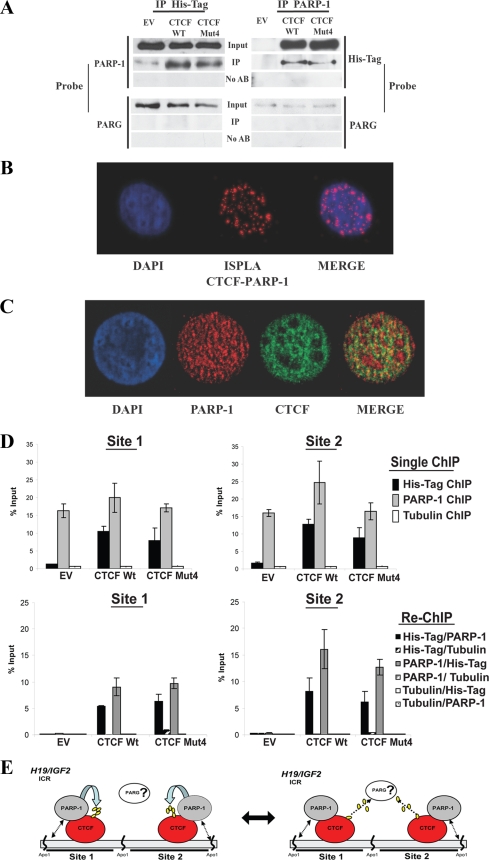
FIG. 5.
CTCF association with PARP-1 provides a mechanism of CTCF functional regulation by PARylation at the H19 ICR and genomewide. (A) Coimmunoprecipitation analysis demonstrating that the association of PARP-1 with CTCF is independent of the CTCF PARylation status. B4 cells (2 × 106) were transfected with either the pCi EV or a vector expressing the CTCF WT or CTCF Mut4 and lysed for IP. The cell lysates were incubated with either the anti-PARP-1 or anti-His tag antibody for 4 h and then with protein A Sepharose Fast Flow for 2 h. The immunoprecipitated proteins were analyzed by Western blotting. The membranes were probed with either the anti-His tag or anti-PARP-1 antibody and sequentially with an anti-PARG antibody. (B) Analysis of CTCF and PARP-1 colocalization by ISPLA. The ISPLA was carried out with MCF7 cells by using both CTCF and PARP-1 antibodies. The red signal observed, as shown in the ISPLA image (center), indicates that CTCF and PARP-1 protein are in close proximity to each other, at a maximum distance of 40 nm or overlapping. DAPI staining and merging of images illustrate the extent of CTCF and PARP-1 nuclear colocalization. (C) Immunofluorescence staining demonstrating CTCF and PARP-1 colocalization. MCF7 cells were fixed and analyzed by dual immunofluorescence staining using both CTCF (green) and PARP-1 (red) antibodies. Nuclei are visualized by DAPI (blue); the merge of the green and red colors in shown in the rightmost image. (D) PARP-1 is associated with both the CTCF WT and CTCF Mut4 at CTCF binding sites 1 and 2 at the H19 ICR in B4 cells, as shown by results from ChIP and re-ChIP assays. B4 cells (5 × 106) were transfected with either the pCi EV or a plasmid expressing the CTCF WT or CTCF Mut4 and cross-linked with formaldehyde. A standard ChIP assay was followed by an additional elution step and re-ChIP to assess the in vivo occupancies of the DNA target sites by PARP-1, the CTCF WT, and CTCF Mut4. An antitubulin antibody was used as the negative control in both the ChIP and re-ChIP experiments. Real-time PCR amplification was carried out using primers situated within site 1 and site 2. The efficiency of ChIP at each of the sites was calculated as a percentage of the starting material (percent input). Expression of the CTCF WT and CTCF Mut4 was assessed by Western blotting (data not shown). (E) Proposed model of the regulation of CTCF functions by PARylation. In this model, the close proximity of CTCF and PARP-1 proteins is suggested to provide a mechanism by which CTCF functional activity may be regulated. CTCF and PARP-1 form a functional complex at the CTCF target sites. (Left) PARP-1 modifies CTCF, thereby modulating CTCF function. (Right) This reaction may be reversed by PARG-1, which may also be associated with the CTCF-PARP-1 complex. Double-headed arrows between PARP-1 and DNA in both the left and right panels indicate that the association between CTCF and PARP-1 may be independent of whether PARP-1 is bound to the DNA.