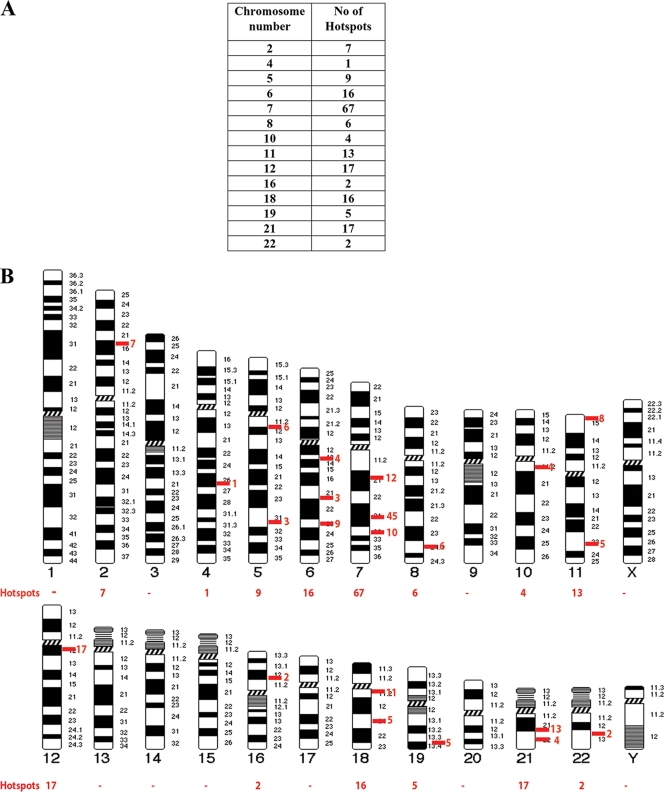
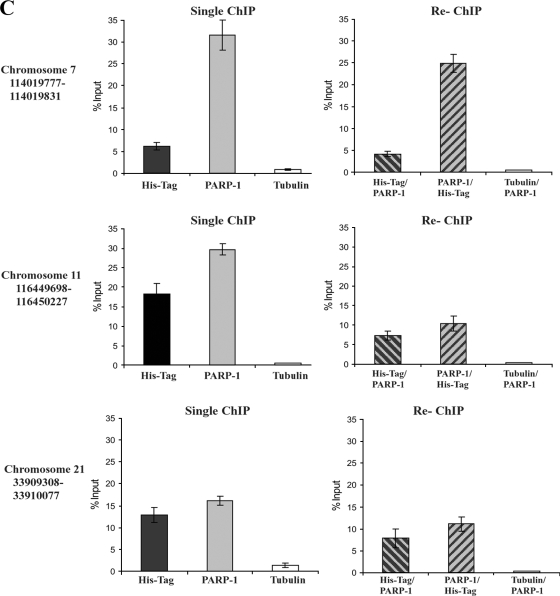
FIG. 6.
Colocalization of CTCF and PARP-1 across the genome. (A) Summary of the microarray data obtained from two independent ChIP-ChIP experiments studying CTCF and PARP-1, deposited in the GEO database. Numerous colocalization sites within human DNA were identified. The table indicates the number of identified CTCF and PARP-1 colocalization hot spots with regard to the specific human chromosomes. (B) Correlated sites (hot spot clusters) of CTCF and PARP-1 colocalization mapped to human chromosomes. The numbers of hot spots on the chromosomes are shown. (C) Validation of the identified CTCF and PARP-1 colocalization sites using three randomly chosen sites positioned on chromosomes 7, 11, and 21. Real-time PCR was performed using a selection of the ChIP samples obtained in the B4 cell experiments (Fig. 5D). The single ChIP samples were analyzed with anti-His tag, anti-PARP-1, and antitubulin antibodies, and re-ChIP samples were analyzed with anti-His tag/anti-PARP-1, anti-PARP-1/anti-His tag, and antitubulin/anti-PARP-1 antibodies. The efficiency of ChIP was calculated as a percentage of the starting material (percent input). At all three chromosomal sites, enrichment is observed, indicating colocalization of CTCF and PARP-1 at these chromosomal positions. (D) Chromosome 11 is shown as an example of a “chromosome region in detail.” The Ensembl genome browser was used to map the sites of CTCF/PARP-1 colocalization. (Top) The chromosomal position of the hot spot cluster comprising five hot spots is shown. (Middle) The region in detail shows genes within the area of CTCF/PARP-1 colocalization. (Bottom) One of the identified colocalization hot spots, hot spot 4, is expanded to reveal the DNA sequence, and a putative CTCF binding site is highlighted. ncRNA, noncoding RNA. (E) Chromosomal positions of the five identified hot spots in a cluster on chromosome 11.